Gene Page: PPP2CA
Summary ?
| GeneID | 5515 |
| Symbol | PPP2CA |
| Synonyms | PP2Ac|PP2CA|PP2Calpha|RP-C |
| Description | protein phosphatase 2 catalytic subunit alpha |
| Reference | MIM:176915|HGNC:HGNC:9299|HPRD:08912| |
| Gene type | protein-coding |
| Map location | 5q31.1 |
| Pascal p-value | 0.42 |
| Sherlock p-value | 0.409 |
| Fetal beta | -0.785 |
| DMG | 1 (# studies) |
| Support | G2Cdb.humanNRC G2Cdb.humanPSD G2Cdb.humanPSP Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0032 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 6.9342 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg23307708 | 5 | 133561976 | PPP2CA | 1.99E-5 | -0.452 | 0.016 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
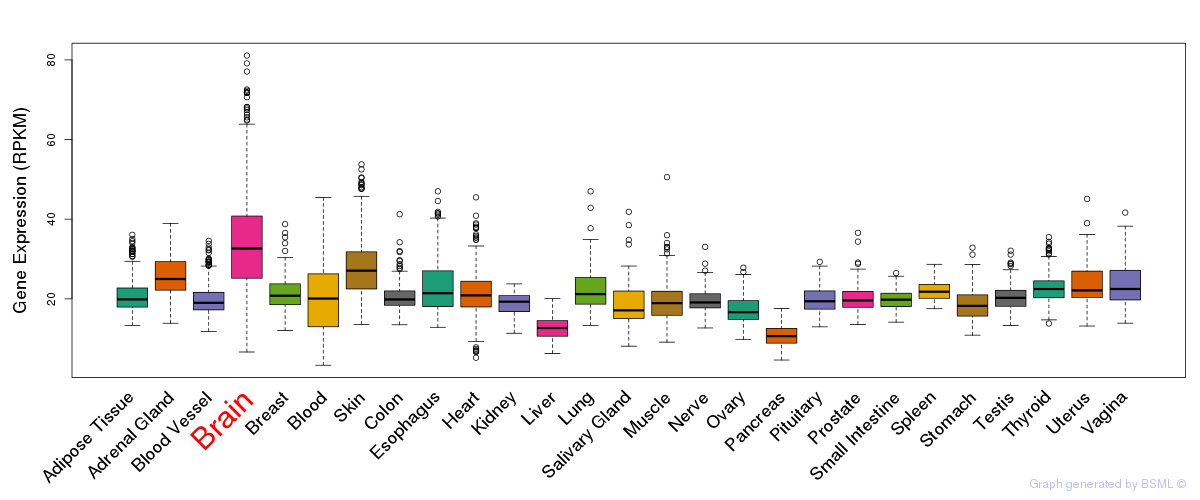
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CHPT1 | 0.80 | 0.78 |
| GYG1 | 0.77 | 0.71 |
| SRI | 0.76 | 0.69 |
| TMEM22 | 0.76 | 0.66 |
| NUDT6 | 0.75 | 0.67 |
| GBAS | 0.75 | 0.69 |
| RAB9A | 0.74 | 0.66 |
| IL17RB | 0.74 | 0.52 |
| HMGCL | 0.74 | 0.66 |
| CYB5R1 | 0.73 | 0.64 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ZNF783 | -0.43 | -0.44 |
| JAG2 | -0.43 | -0.43 |
| AD000671.1 | -0.43 | -0.44 |
| TNKS1BP1 | -0.43 | -0.44 |
| UPF3A | -0.42 | -0.47 |
| BAHCC1 | -0.42 | -0.43 |
| AC010536.2 | -0.42 | -0.43 |
| HIC2 | -0.42 | -0.41 |
| FBN3 | -0.42 | -0.41 |
| SLC26A1 | -0.42 | -0.46 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005506 | iron ion binding | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 9847399 | |
| GO:0004721 | phosphoprotein phosphatase activity | IEA | - | |
| GO:0016787 | hydrolase activity | IEA | - | |
| GO:0030145 | manganese ion binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| GO:0046982 | protein heterodimerization activity | IPI | 9847399 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000188 | inactivation of MAPK activity | NAS | 11007961 | |
| GO:0008380 | RNA splicing | NAS | 11007961 | |
| GO:0006470 | protein amino acid dephosphorylation | TAS | 2849764 |11007961 | |
| GO:0010033 | response to organic substance | NAS | 11007961 | |
| GO:0006275 | regulation of DNA replication | NAS | 11007961 | |
| GO:0006917 | induction of apoptosis | TAS | 11007961 | |
| GO:0006672 | ceramide metabolic process | NAS | 11007961 | |
| GO:0030111 | regulation of Wnt receptor signaling pathway | NAS | 11007961 | |
| GO:0007498 | mesoderm development | IEA | - | |
| GO:0030155 | regulation of cell adhesion | NAS | 11007961 | |
| GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein | NAS | 11007961 | |
| GO:0040008 | regulation of growth | NAS | 11360189 | |
| GO:0019932 | second-messenger-mediated signaling | NAS | 11007961 | |
| GO:0030308 | negative regulation of cell growth | NAS | 11007961 | |
| GO:0045595 | regulation of cell differentiation | NAS | 11360189 | |
| GO:0045449 | regulation of transcription | NAS | 11007961 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000159 | protein phosphatase type 2A complex | TAS | 11007961 | |
| GO:0005829 | cytosol | IEA | - | |
| GO:0005829 | cytosol | TAS | 11007961 | |
| GO:0005625 | soluble fraction | NAS | 11007961 | |
| GO:0005634 | nucleus | NAS | 11007961 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005739 | mitochondrion | NAS | 11007961 | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0015630 | microtubule cytoskeleton | NAS | 11007961 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ARL2 | ARFL2 | ADP-ribosylation factor-like 2 | Co-purification | BioGRID | 12912990 |
| BCL2 | Bcl-2 | B-cell CLL/lymphoma 2 | - | HPRD,BioGRID | 9852076 |
| BEST1 | ARB | BEST | BMD | TU15B | VMD2 | bestrophin 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 12058047 |
| CCNG2 | - | cyclin G2 | Affinity Capture-Western Reconstituted Complex | BioGRID | 11956189 |
| CCT2 | 99D8.1 | CCT-beta | CCTB | MGC142074 | MGC142076 | PRO1633 | TCP-1-beta | chaperonin containing TCP1, subunit 2 (beta) | Affinity Capture-MS | BioGRID | 18782753 |
| CCT3 | CCT-gamma | CCTG | PIG48 | TCP-1-gamma | TRIC5 | chaperonin containing TCP1, subunit 3 (gamma) | Affinity Capture-MS | BioGRID | 18782753 |
| CCT4 | CCT-DELTA | Cctd | MGC126164 | MGC126165 | SRB | chaperonin containing TCP1, subunit 4 (delta) | Affinity Capture-MS | BioGRID | 18782753 |
| CCT5 | CCT-epsilon | CCTE | KIAA0098 | TCP-1-epsilon | chaperonin containing TCP1, subunit 5 (epsilon) | Affinity Capture-MS | BioGRID | 18782753 |
| CCT6A | CCT-zeta | CCT-zeta-1 | CCT6 | Cctz | HTR3 | MGC126214 | MGC126215 | MoDP-2 | TCP-1-zeta | TCP20 | TCPZ | TTCP20 | chaperonin containing TCP1, subunit 6A (zeta 1) | Affinity Capture-MS | BioGRID | 18782753 |
| CCT7 | CCT-ETA | Ccth | MGC110985 | Nip7-1 | TCP-1-eta | chaperonin containing TCP1, subunit 7 (eta) | Affinity Capture-MS | BioGRID | 18782753 |
| CCT8 | C21orf112 | Cctq | D21S246 | KIAA0002 | PRED71 | chaperonin containing TCP1, subunit 8 (theta) | Affinity Capture-MS | BioGRID | 18782753 |
| CDC5L | CEF1 | KIAA0432 | PCDC5RP | dJ319D22.1 | hCDC5 | CDC5 cell division cycle 5-like (S. pombe) | in vitro in vivo Two-hybrid | BioGRID | 10827081 |
| CDK2 | p33(CDK2) | cyclin-dependent kinase 2 | Biochemical Activity Co-purification | BioGRID | 10934208 |
| CDK6 | MGC59692 | PLSTIRE | STQTL11 | cyclin-dependent kinase 6 | Biochemical Activity Co-purification | BioGRID | 10934208 |
| CSNK1A1 | CK1 | HLCDGP1 | PRO2975 | casein kinase 1, alpha 1 | Affinity Capture-MS | BioGRID | 12062430 |
| CTTNBP2 | C7orf8 | CORTBP2 | FLJ34229 | KIAA1758 | MGC104579 | Orf4 | cortactin binding protein 2 | Affinity Capture-MS | BioGRID | 18782753 |
| CTTNBP2NL | DKFZp547A023 | FLJ13278 | CTTNBP2 N-terminal like | Affinity Capture-MS | BioGRID | 18782753 |
| ETF1 | D5S1995 | ERF | ERF1 | MGC111066 | RF1 | SUP45L1 | TB3-1 | eukaryotic translation termination factor 1 | Affinity Capture-Western Co-purification Two-hybrid | BioGRID | 9003791 |
| FAM40A | FLJ14743 | KIAA1761 | MGC148091 | RP4-773N10.1 | family with sequence similarity 40, member A | Affinity Capture-MS | BioGRID | 18782753 |
| FGFR1OP | FOP | FGFR1 oncogene partner | Affinity Capture-MS | BioGRID | 18782753 |
| IGBP1 | ALPHA-4 | IBP1 | immunoglobulin (CD79A) binding protein 1 | Affinity Capture-MS in vivo Two-hybrid | BioGRID | 9647778 |10441131 |16085932 |18782753 |
| IGBP1 | ALPHA-4 | IBP1 | immunoglobulin (CD79A) binding protein 1 | - | HPRD | 9647778 |10441131 |11371618 |
| ISYNA1 | INOS | IPS | Ino1 | inositol-3-phosphate synthase 1 | Two-hybrid | BioGRID | 16169070 |
| MOBKL3 | 2C4D | CGI-95 | MGC12264 | MOB1 | MOB3 | PREI3 | MOB1, Mps One Binder kinase activator-like 3 (yeast) | Affinity Capture-MS | BioGRID | 18782753 |
| MRPS26 | C20orf193 | GI008 | MRP-S13 | MRP-S26 | MRPS13 | NY-BR-87 | RPMS13 | dJ534B8.3 | mitochondrial ribosomal protein S26 | Two-hybrid | BioGRID | 16169070 |
| PDCD10 | CCM3 | MGC1212 | MGC24477 | TFAR15 | programmed cell death 10 | Affinity Capture-MS | BioGRID | 18782753 |
| PPFIA1 | FLJ41337 | FLJ42630 | FLJ43474 | LIP.1 | LIP1 | LIPRIN | MGC26800 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1 | Affinity Capture-MS | BioGRID | 18782753 |
| PPFIA2 | FLJ41378 | MGC132572 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 | Affinity Capture-MS | BioGRID | 18782753 |
| PPFIA3 | KIAA0654 | LPNA3 | MGC126567 | MGC126569 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 | Affinity Capture-MS | BioGRID | 18782753 |
| PPM1B | MGC21657 | PP2C-beta-X | PP2CB | PP2CBETA | PPC2BETAX | protein phosphatase 1B (formerly 2C), magnesium-dependent, beta isoform | Co-purification | BioGRID | 10934208 |
| PPME1 | FLJ22226 | PME-1 | protein phosphatase methylesterase 1 | Affinity Capture-MS | BioGRID | 18782753 |
| PPP2R1A | MGC786 | PR65A | protein phosphatase 2 (formerly 2A), regulatory subunit A, alpha isoform | Affinity Capture-MS Affinity Capture-Western Reconstituted Complex | BioGRID | 12370081 |18782753 |
| PPP2R1A | MGC786 | PR65A | protein phosphatase 2 (formerly 2A), regulatory subunit A, alpha isoform | A-alpha subunit interacts with C-alpha subunit of PP2A. | BIND | 12370081 |
| PPP2R1B | MGC26454 | PR65B | protein phosphatase 2 (formerly 2A), regulatory subunit A, beta isoform | Affinity Capture-MS Affinity Capture-Western Reconstituted Complex | BioGRID | 12370081 |18782753 |
| PPP2R1B | MGC26454 | PR65B | protein phosphatase 2 (formerly 2A), regulatory subunit A, beta isoform | A-beta subunit interacts with C-alpha subunit of PP2A. This interaction was modeled on a demonstrated interaction between human A-beta subunit and rabbit C-alpha subunit. | BIND | 11313937 |
| PPP2R2A | B55-ALPHA | B55A | FLJ26613 | MGC52248 | PR52A | PR55A | protein phosphatase 2 (formerly 2A), regulatory subunit B, alpha isoform | - | HPRD,BioGRID | 1328247 |
| PPP2R2C | B55-GAMMA | IMYPNO | IMYPNO1 | MGC33570 | PR52 | PR55G | protein phosphatase 2 (formerly 2A), regulatory subunit B, gamma isoform | Affinity Capture-MS | BioGRID | 18782753 |
| PPP2R2D | MDS026 | protein phosphatase 2, regulatory subunit B, delta isoform | Affinity Capture-MS | BioGRID | 18782753 |
| PPP2R3A | PPP2R3 | PR130 | PR72 | protein phosphatase 2 (formerly 2A), regulatory subunit B'', alpha | Two-hybrid | BioGRID | 10629059 |
| PPP2R3B | NY-REN-8 | PPP2R3L | PPP2R3LY | PR48 | protein phosphatase 2 (formerly 2A), regulatory subunit B'', beta | Affinity Capture-MS Two-hybrid | BioGRID | 10629059 |18782753 |
| PPP2R5A | B56A | MGC131915 | PR61A | protein phosphatase 2, regulatory subunit B', alpha isoform | Affinity Capture-MS | BioGRID | 18782753 |
| PPP2R5B | B56B | FLJ35411 | PR61B | protein phosphatase 2, regulatory subunit B', beta isoform | Affinity Capture-MS | BioGRID | 18782753 |
| PPP2R5C | B56G | MGC23064 | PR61G | protein phosphatase 2, regulatory subunit B', gamma isoform | Affinity Capture-MS Affinity Capture-Western | BioGRID | 10675325 |18782753 |
| PPP2R5D | B56D | MGC2134 | MGC8949 | protein phosphatase 2, regulatory subunit B', delta isoform | Affinity Capture-MS | BioGRID | 18782753 |
| PPP2R5E | - | protein phosphatase 2, regulatory subunit B', epsilon isoform | Affinity Capture-MS in vivo | BioGRID | 8703017 |18782753 |
| PRKCD | MAY1 | MGC49908 | PKCD | nPKC-delta | protein kinase C, delta | Biochemical Activity | BioGRID | 11959144 |
| PTN | HARP | HBGF8 | HBNF | NEGF1 | pleiotrophin | Two-hybrid | BioGRID | 16169070 |
| PXN | FLJ16691 | paxillin | Affinity Capture-Western | BioGRID | 10675325 |
| RELA | MGC131774 | NFKB3 | p65 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | Reconstituted Complex | BioGRID | 11591705 |
| RP5-1000E10.4 | DKFZp686A0768 | FLJ21168 | SIKE | suppressor of IKK epsilon | Affinity Capture-MS | BioGRID | 18782753 |
| RP6-213H19.1 | MASK | MST4 | serine/threonine protein kinase MST4 | Affinity Capture-MS | BioGRID | 18782753 |
| RPS6KB1 | PS6K | S6K | S6K1 | STK14A | p70(S6K)-alpha | p70-S6K | p70-alpha | ribosomal protein S6 kinase, 70kDa, polypeptide 1 | - | HPRD | 11438723 |
| STK24 | MST-3 | MST3 | MST3B | STE20 | STK3 | serine/threonine kinase 24 (STE20 homolog, yeast) | Affinity Capture-MS | BioGRID | 18782753 |
| STK25 | DKFZp686J1430 | SOK1 | YSK1 | serine/threonine kinase 25 (STE20 homolog, yeast) | Affinity Capture-MS | BioGRID | 18782753 |
| STRN | MGC125642 | SG2NA | striatin, calmodulin binding protein | Affinity Capture-MS | BioGRID | 18782753 |
| STRN | MGC125642 | SG2NA | striatin, calmodulin binding protein | - | HPRD | 10681496 |
| STRN3 | SG2NA | striatin, calmodulin binding protein 3 | Affinity Capture-MS | BioGRID | 18782753 |
| STRN4 | FLJ35594 | ZIN | zinedin | striatin, calmodulin binding protein 4 | Affinity Capture-MS | BioGRID | 18782753 |
| TBCD | KIAA0988 | tubulin folding cofactor D | Affinity Capture-Western | BioGRID | 12912990 |
| TCP1 | CCT-alpha | CCT1 | CCTa | D6S230E | TCP-1-alpha | t-complex 1 | Affinity Capture-MS | BioGRID | 18782753 |
| TLX1 | HOX11 | MGC163402 | TCL3 | T-cell leukemia homeobox 1 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 9009195 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | p53 interacts with PPP2CA. This interaction is modeled on a demonstrated interaction between p53 from an unspecified source and human PP2AC. | BIND | 12556559 |
| TRAF3IP3 | DJ434O14.3 | FLJ44151 | MGC117354 | MGC163289 | T3JAM | TRAF3 interacting protein 3 | Affinity Capture-MS | BioGRID | 18782753 |
| TRIP13 | 16E1BP | thyroid hormone receptor interactor 13 | Two-hybrid | BioGRID | 16189514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG OOCYTE MEIOSIS | 114 | 79 | All SZGR 2.0 genes in this pathway |
| KEGG WNT SIGNALING PATHWAY | 151 | 112 | All SZGR 2.0 genes in this pathway |
| KEGG TGF BETA SIGNALING PATHWAY | 86 | 64 | All SZGR 2.0 genes in this pathway |
| KEGG TIGHT JUNCTION | 134 | 86 | All SZGR 2.0 genes in this pathway |
| KEGG LONG TERM DEPRESSION | 70 | 53 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AKAP95 PATHWAY | 12 | 9 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AKT PATHWAY | 22 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CTCF PATHWAY | 23 | 18 | All SZGR 2.0 genes in this pathway |
| BIOCARTA P35ALZHEIMERS PATHWAY | 11 | 9 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ERK PATHWAY | 28 | 22 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GSK3 PATHWAY | 27 | 26 | All SZGR 2.0 genes in this pathway |
| BIOCARTA KERATINOCYTE PATHWAY | 46 | 38 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MTOR PATHWAY | 23 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AKAPCENTROSOME PATHWAY | 15 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CHREBP2 PATHWAY | 42 | 35 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CK1 PATHWAY | 17 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IGF1MTOR PATHWAY | 20 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TEL PATHWAY | 18 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA WNT PATHWAY | 26 | 24 | All SZGR 2.0 genes in this pathway |
| WNT SIGNALING | 89 | 71 | All SZGR 2.0 genes in this pathway |
| PID ATR PATHWAY | 39 | 25 | All SZGR 2.0 genes in this pathway |
| PID PLK1 PATHWAY | 46 | 25 | All SZGR 2.0 genes in this pathway |
| PID MYC PATHWAY | 25 | 22 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 DOWNSTREAM PATHWAY | 105 | 78 | All SZGR 2.0 genes in this pathway |
| PID PDGFRB PATHWAY | 129 | 103 | All SZGR 2.0 genes in this pathway |
| PID IL8 CXCR2 PATHWAY | 34 | 26 | All SZGR 2.0 genes in this pathway |
| PID P53 REGULATION PATHWAY | 59 | 50 | All SZGR 2.0 genes in this pathway |
| PID RB 1PATHWAY | 65 | 46 | All SZGR 2.0 genes in this pathway |
| PID TGFBR PATHWAY | 55 | 38 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY WNT | 65 | 41 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME CTNNB1 PHOSPHORYLATION CASCADE | 16 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME SPRY REGULATION OF FGF SIGNALING | 14 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME TRIF MEDIATED TLR3 SIGNALING | 74 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME NEGATIVE REGULATION OF FGFR SIGNALING | 37 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | 137 | 105 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR IN DISEASE | 127 | 88 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | 24 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME ERKS ARE INACTIVATED | 12 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME ERK MAPK TARGETS | 21 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME GLYCOLYSIS | 29 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRATION OF ENERGY METABOLISM | 120 | 84 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME OPIOID SIGNALLING | 78 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME DARPP 32 EVENTS | 25 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME G1 PHASE | 38 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME G1 S TRANSITION | 112 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME COSTIMULATION BY THE CD28 FAMILY | 63 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME CTLA4 INHIBITORY SIGNALING | 21 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF MRNA | 284 | 128 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF RNA | 330 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC G1 G1 S PHASES | 137 | 79 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC M M G1 PHASES | 172 | 98 | All SZGR 2.0 genes in this pathway |
| REACTOME MAP KINASE ACTIVATION IN TLR CASCADE | 50 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME MAPK TARGETS NUCLEAR EVENTS MEDIATED BY MAP KINASES | 30 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET HOMEOSTASIS | 78 | 49 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET SENSITIZATION BY LDL | 16 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAF6 MEDIATED INDUCTION OF NFKB AND MAP KINASES UPON TLR7 8 OR 9 ACTIVATION | 77 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME NFKB AND MAP KINASES ACTIVATION MEDIATED BY TLR4 SIGNALING REPERTOIRE | 72 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | 13 | 6 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPLICATION | 192 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME E2F MEDIATED REGULATION OF DNA REPLICATION | 35 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF CARBOHYDRATES | 247 | 154 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | 83 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC PROMETAPHASE | 87 | 51 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATED TLR4 SIGNALLING | 93 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME TOLL RECEPTOR CASCADES | 118 | 84 | All SZGR 2.0 genes in this pathway |
| REACTOME GLUCOSE METABOLISM | 69 | 42 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | 176 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR | 112 | 80 | All SZGR 2.0 genes in this pathway |
| WATANABE RECTAL CANCER RADIOTHERAPY RESPONSIVE DN | 92 | 51 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER DN | 406 | 230 | All SZGR 2.0 genes in this pathway |
| BIDUS METASTASIS UP | 214 | 134 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| COLLIS PRKDC REGULATORS | 15 | 10 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPLICATION GENES | 147 | 87 | All SZGR 2.0 genes in this pathway |
| SMITH TERT TARGETS UP | 145 | 91 | All SZGR 2.0 genes in this pathway |
| LU IL4 SIGNALING | 94 | 56 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| NATSUME RESPONSE TO INTERFERON BETA DN | 52 | 33 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C1 | 72 | 45 | All SZGR 2.0 genes in this pathway |
| JIANG AGING CEREBRAL CORTEX DN | 54 | 43 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY GAMMA RADIATION | 81 | 59 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE DN | 195 | 135 | All SZGR 2.0 genes in this pathway |
| DAIRKEE CANCER PRONE RESPONSE BPA E2 | 118 | 65 | All SZGR 2.0 genes in this pathway |
| HOFFMANN LARGE TO SMALL PRE BII LYMPHOCYTE UP | 163 | 102 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 4HR DN | 254 | 158 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE M G1 | 148 | 95 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 3 UP | 170 | 97 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 6 UP | 140 | 81 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 7 UP | 118 | 68 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| IKEDA MIR133 TARGETS UP | 43 | 27 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-125/351 | 476 | 482 | 1A | hsa-miR-125bbrain | UCCCUGAGACCCUAACUUGUGA |
| hsa-miR-125abrain | UCCCUGAGACCCUUUAACCUGUG | ||||
| miR-133 | 123 | 129 | 1A | hsa-miR-133a | UUGGUCCCCUUCAACCAGCUGU |
| hsa-miR-133b | UUGGUCCCCUUCAACCAGCUA | ||||
| miR-139 | 365 | 372 | 1A,m8 | hsa-miR-139brain | UCUACAGUGCACGUGUCU |
| miR-141/200a | 645 | 651 | m8 | hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG |
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| miR-17-5p/20/93.mr/106/519.d | 180 | 186 | 1A | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-183 | 135 | 142 | 1A,m8 | hsa-miR-183 | UAUGGCACUGGUAGAAUUCACUG |
| miR-188 | 478 | 484 | 1A | hsa-miR-188 | CAUCCCUUGCAUGGUGGAGGGU |
| miR-200bc/429 | 19 | 25 | m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-219 | 388 | 394 | 1A | hsa-miR-219brain | UGAUUGUCCAAACGCAAUUCU |
| miR-29 | 718 | 724 | m8 | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
| miR-409-3p | 97 | 104 | 1A,m8 | hsa-miR-409-3p | CGAAUGUUGCUCGGUGAACCCCU |
| miR-452 | 384 | 390 | 1A | hsa-miR-452 | UGUUUGCAGAGGAAACUGAGAC |
| miR-495 | 557 | 563 | 1A | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.