Gene Page: PRKDC
Summary ?
| GeneID | 5591 |
| Symbol | PRKDC |
| Synonyms | DNA-PKcs|DNAPK|DNPK1|HYRC|HYRC1|IMD26|XRCC7|p350 |
| Description | protein kinase, DNA-activated, catalytic polypeptide |
| Reference | MIM:600899|HGNC:HGNC:9413|Ensembl:ENSG00000253729|HPRD:02941|Vega:OTTHUMG00000164239 |
| Gene type | protein-coding |
| Map location | 8q11 |
| Pascal p-value | 0.305 |
| Sherlock p-value | 0.681 |
| Fetal beta | 0.866 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 1 |
| DNM:McCarthy_2014 | Whole Exome Sequencing analysis | Whole exome sequencing of 57 trios with sporadic or familial schizophrenia. | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0381 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PRKDC | chr8 | 48868449 | UNKNOWN | NA | synonymous SNV | Schizophrenia | DNM:McCarthy_2014 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg16104446 | 8 | 48873951 | PRKDC | 0.005 | -7.73 | DMG:vanEijk_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7132043 | chr12 | 80968399 | PRKDC | 5591 | 0.07 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
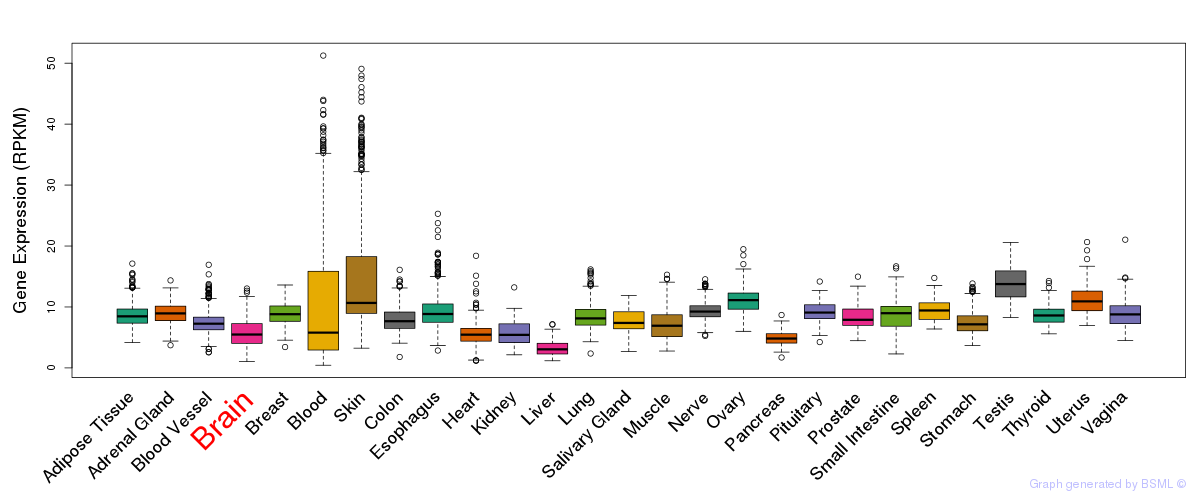
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| LRRC24 | 0.79 | 0.74 |
| LRFN4 | 0.75 | 0.75 |
| COMTD1 | 0.74 | 0.68 |
| AC006273.1 | 0.73 | 0.72 |
| ZNF768 | 0.73 | 0.77 |
| FAM171A2 | 0.73 | 0.73 |
| MACROD1 | 0.73 | 0.69 |
| GPR172A | 0.73 | 0.74 |
| MFSD10 | 0.73 | 0.74 |
| AC009133.1 | 0.72 | 0.69 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| S100B | -0.51 | -0.60 |
| AF347015.27 | -0.49 | -0.57 |
| C5orf53 | -0.49 | -0.55 |
| ACOT13 | -0.48 | -0.53 |
| PIR | -0.48 | -0.56 |
| COPZ2 | -0.47 | -0.54 |
| AF347015.33 | -0.47 | -0.56 |
| LACTB2 | -0.47 | -0.53 |
| CSRP1 | -0.47 | -0.53 |
| SLCO1A2 | -0.47 | -0.58 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003677 | DNA binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 9442054 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004677 | DNA-dependent protein kinase activity | IDA | 15194694 | |
| GO:0016740 | transferase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007420 | brain development | IEA | Brain (GO term level: 7) | - |
| GO:0001756 | somitogenesis | IEA | - | |
| GO:0000723 | telomere maintenance | IEA | - | |
| GO:0002328 | pro-B cell differentiation | IEA | - | |
| GO:0006303 | double-strand break repair via nonhomologous end joining | IEA | - | |
| GO:0006310 | DNA recombination | IEA | - | |
| GO:0006464 | protein modification process | TAS | 7671312 | |
| GO:0010332 | response to gamma radiation | IEA | - | |
| GO:0007507 | heart development | IEA | - | |
| GO:0006915 | apoptosis | IEA | - | |
| GO:0018105 | peptidyl-serine phosphorylation | IDA | 15194694 | |
| GO:0033077 | T cell differentiation in the thymus | IEA | - | |
| GO:0033153 | T cell receptor V(D)J recombination | IEA | - | |
| GO:0033152 | immunoglobulin V(D)J recombination | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005654 | nucleoplasm | EXP | 12023295 |12065431 | |
| GO:0005958 | DNA-dependent protein kinase complex | IDA | 15194694 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABL1 | ABL | JTK7 | bcr/abl | c-ABL | p150 | v-abl | c-abl oncogene 1, receptor tyrosine kinase | - | HPRD,BioGRID | 9312071 |
| AKT1 | AKT | MGC99656 | PKB | PKB-ALPHA | PRKBA | RAC | RAC-ALPHA | v-akt murine thymoma viral oncogene homolog 1 | DNA-PKcs interacts with Akt1. | BIND | 15678105 |
| AKT2 | PKBB | PKBBETA | PRKBB | RAC-BETA | v-akt murine thymoma viral oncogene homolog 2 | Biochemical Activity | BioGRID | 16221682 |
| AP1B1 | ADTB1 | AP105A | BAM22 | CLAPB2 | adaptor-related protein complex 1, beta 1 subunit | Protein-peptide | BioGRID | 10608806 |
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | Affinity Capture-MS | BioGRID | 15640154 |
| ATM | AT1 | ATA | ATC | ATD | ATDC | ATE | DKFZp781A0353 | MGC74674 | TEL1 | TELO1 | ataxia telangiectasia mutated | - | HPRD,BioGRID | 10464290 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | Protein-peptide | BioGRID | 10608806 |
| C1D | MGC12261 | MGC14659 | SUNCOR | nuclear DNA-binding protein | - | HPRD,BioGRID | 9679063 |
| CHEK1 | CHK1 | CHK1 checkpoint homolog (S. pombe) | Protein-peptide Reconstituted Complex | BioGRID | 10608806 |12756247 |
| CHUK | IKBKA | IKK-alpha | IKK1 | IKKA | NFKBIKA | TCF16 | conserved helix-loop-helix ubiquitous kinase | - | HPRD,BioGRID | 9632806 |
| CIB1 | CIB | KIP | KIP1 | SIP2-28 | calcium and integrin binding 1 (calmyrin) | - | HPRD,BioGRID | 9372844 |
| DCLRE1C | A-SCID | DCLREC1C | FLJ11360 | FLJ36438 | RS-SCID | SCIDA | SNM1C | DNA cross-link repair 1C (PSO2 homolog, S. cerevisiae) | Affinity Capture-Western Biochemical Activity Reconstituted Complex | BioGRID | 11955432 |
| E4F1 | E4F | MGC99614 | E4F transcription factor 1 | Protein-peptide | BioGRID | 10608806 |
| EIF2S2 | DKFZp686L18198 | EIF2 | EIF2B | EIF2beta | MGC8508 | eukaryotic translation initiation factor 2, subunit 2 beta, 38kDa | - | HPRD,BioGRID | 9442054 |
| HOXC4 | HOX3 | HOX3E | cp19 | homeobox C4 | - | HPRD | 11279128 |
| HSF1 | HSTF1 | heat shock transcription factor 1 | in vitro | BioGRID | 9325337 |
| IKBKB | FLJ40509 | IKK-beta | IKK2 | IKKB | MGC131801 | NFKBIKB | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta | Biochemical Activity | BioGRID | 9632806 |
| ILF2 | MGC8391 | NF45 | PRO3063 | interleukin enhancer binding factor 2, 45kDa | - | HPRD,BioGRID | 9442054 |
| ILF3 | CBTF | DRBF | DRBP76 | MMP4 | MPHOSPH4 | MPP4 | NF-AT-90 | NF110 | NF90 | NFAR | NFAR-1 | NFAR2 | TCP110 | TCP80 | interleukin enhancer binding factor 3, 90kDa | - | HPRD,BioGRID | 9442054 |
| KAT2A | GCN5 | GCN5L2 | MGC102791 | PCAF-b | hGCN5 | K(lysine) acetyltransferase 2A | Biochemical Activity | BioGRID | 9488450 |
| LIG4 | - | ligase IV, DNA, ATP-dependent | Protein-peptide | BioGRID | 10608806 |
| LYN | FLJ26625 | JTK8 | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog | - | HPRD,BioGRID | 9748231 |
| MAPK8 | JNK | JNK1 | JNK1A2 | JNK21B1/2 | PRKM8 | SAPK1 | mitogen-activated protein kinase 8 | - | HPRD,BioGRID | 11749722 |
| MAPK9 | JNK-55 | JNK2 | JNK2A | JNK2ALPHA | JNK2B | JNK2BETA | PRKM9 | SAPK | p54a | p54aSAPK | mitogen-activated protein kinase 9 | Biochemical Activity | BioGRID | 11749722 |
| MRE11A | ATLD | HNGS1 | MRE11 | MRE11B | MRE11 meiotic recombination 11 homolog A (S. cerevisiae) | Protein-peptide | BioGRID | 10608806 |
| NBN | AT-V1 | AT-V2 | ATV | FLJ10155 | MGC87362 | NBS | NBS1 | P95 | nibrin | NBS1 interacts with PRKDC (DNA-PKc). | BIND | 15758953 |
| NCF1 | FLJ79451 | NCF1A | NOXO2 | SH3PXD1A | p47phox | neutrophil cytosolic factor 1 | Biochemical Activity | BioGRID | 9914162 |
| NCF2 | FLJ93058 | NCF-2 | NOXA2 | P67-PHOX | P67PHOX | neutrophil cytosolic factor 2 | Biochemical Activity | BioGRID | 9914162 |
| NCF4 | MGC3810 | NCF | P40PHOX | SH3PXD4 | neutrophil cytosolic factor 4, 40kDa | Biochemical Activity | BioGRID | 9914162 |
| NCOA6 | AIB3 | ANTP | ASC2 | HOX1.1 | HOXA7 | KIAA0181 | NRC | PRIP | RAP250 | TRBP | nuclear receptor coactivator 6 | Affinity Capture-MS Biochemical Activity | BioGRID | 10823961 |
| PARP1 | ADPRT | ADPRT1 | PARP | PARP-1 | PPOL | pADPRT-1 | poly (ADP-ribose) polymerase 1 | - | HPRD | 9398855 |
| PCNA | MGC8367 | proliferating cell nuclear antigen | - | HPRD,BioGRID | 12171929 |
| PCNA | MGC8367 | proliferating cell nuclear antigen | PRKDC is a PCNA-binding protein | BIND | 12171929 |
| PRKCD | MAY1 | MGC49908 | PKCD | nPKC-delta | protein kinase C, delta | - | HPRD | 9774685 |
| PTS | FLJ97081 | PTPS | 6-pyruvoyltetrahydropterin synthase | Protein-peptide | BioGRID | 10608806 |
| RAD17 | CCYC | FLJ41520 | HRAD17 | R24L | RAD17SP | RAD24 | RAD17 homolog (S. pombe) | Protein-peptide | BioGRID | 10608806 |
| RBBP8 | CTIP | RIM | retinoblastoma binding protein 8 | Protein-peptide | BioGRID | 10608806 |
| RPA1 | HSSB | REPA1 | RF-A | RP-A | RPA70 | replication protein A1, 70kDa | - | HPRD,BioGRID | 10064605 |
| RPA2 | REPA2 | RPA32 | replication protein A2, 32kDa | DNA-PKcs interacts with RPA2. | BIND | 11000264 |
| RPA2 | REPA2 | RPA32 | replication protein A2, 32kDa | - | HPRD,BioGRID | 10064605 |11731442 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | - | HPRD | 10470151 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | Biochemical Activity Protein-peptide | BioGRID | 9679063 |10608806 |12756247 |
| TRADD | Hs.89862 | MGC11078 | TNFRSF1A-associated via death domain | - | HPRD | 14743216 |
| WRN | DKFZp686C2056 | RECQ3 | RECQL2 | RECQL3 | Werner syndrome | - | HPRD,BioGRID | 11889123 |
| XRCC4 | - | X-ray repair complementing defective repair in Chinese hamster cells 4 | - | HPRD | 9430729 |
| XRCC5 | FLJ39089 | KARP-1 | KARP1 | KU80 | KUB2 | Ku86 | NFIV | X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining) | - | HPRD | 10207052 |10446239 |
| XRCC5 | FLJ39089 | KARP-1 | KARP1 | KU80 | KUB2 | Ku86 | NFIV | X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining) | XRCC5 (Ku80) interacts with PRKDC (DNA-PKc). | BIND | 15758953 |
| XRCC5 | FLJ39089 | KARP-1 | KARP1 | KU80 | KUB2 | Ku86 | NFIV | X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining) | Affinity Capture-Western in vitro in vivo Reconstituted Complex | BioGRID | 9312071 |10446239 |12393188 |
| XRCC6 | CTC75 | CTCBF | G22P1 | KU70 | ML8 | TLAA | X-ray repair complementing defective repair in Chinese hamster cells 6 | - | HPRD | 9312071 |10446239 |11279128 |
| XRCC6 | CTC75 | CTCBF | G22P1 | KU70 | ML8 | TLAA | X-ray repair complementing defective repair in Chinese hamster cells 6 | Affinity Capture-Western in vitro in vivo | BioGRID | 10064605 |10446239 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NON HOMOLOGOUS END JOINING | 14 | 8 | All SZGR 2.0 genes in this pathway |
| KEGG CELL CYCLE | 128 | 84 | All SZGR 2.0 genes in this pathway |
| BIOCARTA G2 PATHWAY | 24 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA FAS PATHWAY | 30 | 22 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HIVNEF PATHWAY | 58 | 43 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TNFR1 PATHWAY | 29 | 21 | All SZGR 2.0 genes in this pathway |
| PID DNA PK PATHWAY | 16 | 10 | All SZGR 2.0 genes in this pathway |
| PID AR PATHWAY | 61 | 46 | All SZGR 2.0 genes in this pathway |
| PID PI3KCI AKT PATHWAY | 35 | 30 | All SZGR 2.0 genes in this pathway |
| PID BARD1 PATHWAY | 29 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME DOUBLE STRAND BREAK REPAIR | 24 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPAIR | 112 | 59 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA UP | 294 | 178 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA PROGENITOR DN | 66 | 42 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA UP | 305 | 185 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL UP | 133 | 78 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS UP | 290 | 177 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY DN | 362 | 238 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| BIDUS METASTASIS UP | 214 | 134 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED UP | 633 | 376 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| OUELLET OVARIAN CANCER INVASIVE VS LMP UP | 117 | 85 | All SZGR 2.0 genes in this pathway |
| MISSIAGLIA REGULATED BY METHYLATION DN | 122 | 67 | All SZGR 2.0 genes in this pathway |
| WAKASUGI HAVE ZNF143 BINDING SITES | 58 | 33 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER CLUSTER 2 | 33 | 17 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| PUJANA BREAST CANCER LIT INT NETWORK | 101 | 73 | All SZGR 2.0 genes in this pathway |
| PUJANA XPRSS INT NETWORK | 168 | 103 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA CENTERED NETWORK | 117 | 72 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST DN | 309 | 206 | All SZGR 2.0 genes in this pathway |
| FERNANDEZ BOUND BY MYC | 182 | 116 | All SZGR 2.0 genes in this pathway |
| REN BOUND BY E2F | 61 | 40 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA SUBGROUPS | 30 | 20 | All SZGR 2.0 genes in this pathway |
| MOREAUX B LYMPHOCYTE MATURATION BY TACI DN | 73 | 45 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI DN | 172 | 107 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR | 532 | 309 | All SZGR 2.0 genes in this pathway |
| LEE AGING NEOCORTEX DN | 80 | 49 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| FUJII YBX1 TARGETS DN | 202 | 132 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN DN | 249 | 165 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE DN | 315 | 197 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL OPTIMAL DEBULKING | 246 | 152 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| GRADE COLON AND RECTAL CANCER UP | 285 | 167 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| FIRESTEIN PROLIFERATION | 175 | 125 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6 | 456 | 285 | All SZGR 2.0 genes in this pathway |
| DING LUNG CANCER BY MUTATION RATE | 20 | 18 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G123 UP | 45 | 21 | All SZGR 2.0 genes in this pathway |
| FOURNIER ACINAR DEVELOPMENT LATE 2 | 277 | 172 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| WIERENGA PML INTERACTOME | 42 | 23 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| ABRAMSON INTERACT WITH AIRE | 45 | 33 | All SZGR 2.0 genes in this pathway |