Gene Page: ZBTB16
Summary ?
| GeneID | 7704 |
| Symbol | ZBTB16 |
| Synonyms | PLZF|ZNF145 |
| Description | zinc finger and BTB domain containing 16 |
| Reference | MIM:176797|HGNC:HGNC:12930|Ensembl:ENSG00000109906|HPRD:11762|Vega:OTTHUMG00000168243 |
| Gene type | protein-coding |
| Map location | 11q23.1 |
| Pascal p-value | 0.733 |
| Sherlock p-value | 0.64 |
| Fetal beta | -2.017 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.006 | |
| Expression | Meta-analysis of gene expression | P value: 2.137 | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg07673133 | 11 | 113930011 | ZBTB16 | 1.22E-8 | -0.015 | 4.99E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
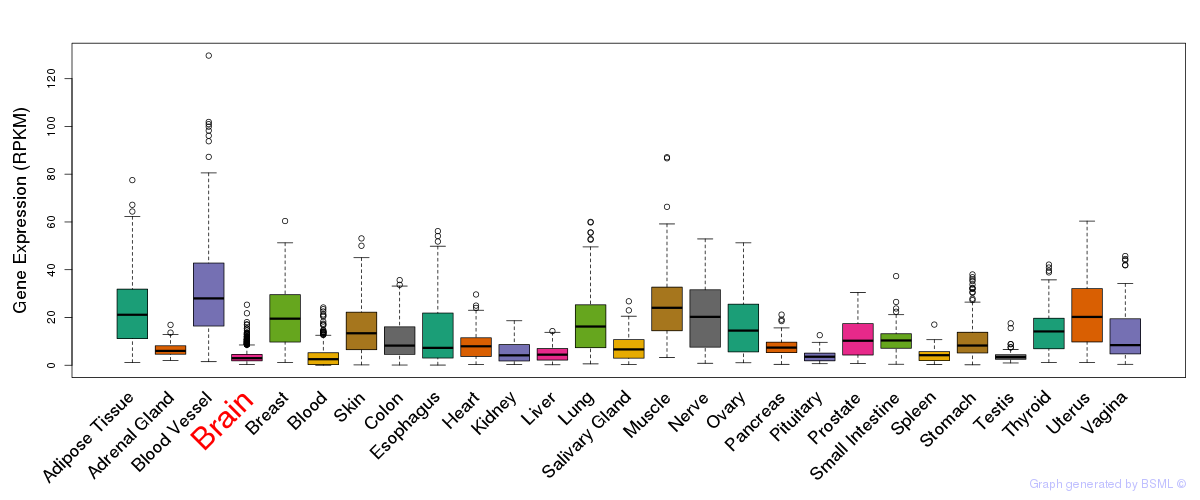
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DDX5 | 0.96 | 0.94 |
| ZNF285B | 0.95 | 0.91 |
| UIMC1 | 0.95 | 0.88 |
| C1orf26 | 0.95 | 0.93 |
| CEP57 | 0.95 | 0.93 |
| HSF2 | 0.94 | 0.93 |
| ZCCHC7 | 0.94 | 0.94 |
| ZNF586 | 0.94 | 0.88 |
| WTAP | 0.94 | 0.94 |
| TEX10 | 0.94 | 0.91 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HLA-F | -0.71 | -0.80 |
| AIFM3 | -0.70 | -0.79 |
| AF347015.31 | -0.69 | -0.87 |
| AF347015.27 | -0.69 | -0.86 |
| MT-CO2 | -0.68 | -0.87 |
| FBXO2 | -0.68 | -0.70 |
| PTH1R | -0.68 | -0.74 |
| C5orf53 | -0.67 | -0.72 |
| IFI27 | -0.67 | -0.86 |
| MT-CYB | -0.67 | -0.86 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003677 | DNA binding | NAS | 9294197 | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0016566 | specific transcriptional repressor activity | IDA | 12802276 | |
| GO:0042803 | protein homodimerization activity | IDA | 8622986 | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007417 | central nervous system development | ISS | Brain (GO term level: 6) | 8541544 |
| GO:0001823 | mesonephros development | ISS | 8541544 | |
| GO:0006350 | transcription | IEA | - | |
| GO:0006511 | ubiquitin-dependent protein catabolic process | IEA | - | |
| GO:0006915 | apoptosis | NAS | 9294197 | |
| GO:0045638 | negative regulation of myeloid cell differentiation | ISS | 8541544 | |
| GO:0045892 | negative regulation of transcription, DNA-dependent | IDA | 12802276 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005634 | nucleus | IDA | 9294197 | |
| GO:0005730 | nucleolus | IDA | 18029348 | |
| GO:0016607 | nuclear speck | IDA | 8541544 | |
| GO:0016605 | PML body | IDA | 9294197 | |
| GO:0017053 | transcriptional repressor complex | IDA | 12802276 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AGTR1 | AG2S | AGTR1A | AGTR1B | AT1 | AT1B | AT1R | AT2R1 | AT2R1A | AT2R1B | HAT1R | angiotensin II receptor, type 1 | Reconstituted Complex Two-hybrid | BioGRID | 14657020 |
| AGTR2 | AT2 | ATGR2 | MRX88 | angiotensin II receptor, type 2 | - | HPRD | 14657020 |
| ANAPC5 | APC5 | anaphase promoting complex subunit 5 | Two-hybrid | BioGRID | 16169070 |
| BCL6 | BCL5 | BCL6A | LAZ3 | ZBTB27 | ZNF51 | B-cell CLL/lymphoma 6 | - | HPRD,BioGRID | 11175338 |
| BMI1 | MGC12685 | PCGF4 | RNF51 | BMI1 polycomb ring finger oncogene | - | HPRD,BioGRID | 12408802 |
| CCDC109B | FLJ20647 | coiled-coil domain containing 109B | Two-hybrid | BioGRID | 16169070 |
| CCDC130 | MGC10471 | coiled-coil domain containing 130 | Two-hybrid | BioGRID | 16169070 |
| CCDC85B | DIPA | coiled-coil domain containing 85B | Two-hybrid | BioGRID | 16189514 |
| CD81 | S5.7 | TAPA1 | TSPAN28 | CD81 molecule | Two-hybrid | BioGRID | 16169070 |
| CDC2 | CDC28A | CDK1 | DKFZp686L20222 | MGC111195 | cell division cycle 2, G1 to S and G2 to M | - | HPRD,BioGRID | 10497277 |
| CEP70 | BITE | FLJ13036 | centrosomal protein 70kDa | Two-hybrid | BioGRID | 16189514 |
| CEP72 | FLJ10565 | KIAA1519 | MGC5307 | centrosomal protein 72kDa | Two-hybrid | BioGRID | 16189514 |
| CIDEA | CIDE-A | cell death-inducing DFFA-like effector a | Two-hybrid | BioGRID | 16169070 |
| COQ6 | CGI-10 | coenzyme Q6 homolog, monooxygenase (S. cerevisiae) | Two-hybrid | BioGRID | 16169070 |
| DLST | DLTS | dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) | Two-hybrid | BioGRID | 16169070 |
| DPM1 | CDGIE | MPDS | dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit | Two-hybrid | BioGRID | 16169070 |
| EIF2S2 | DKFZp686L18198 | EIF2 | EIF2B | EIF2beta | MGC8508 | eukaryotic translation initiation factor 2, subunit 2 beta, 38kDa | Two-hybrid | BioGRID | 16169070 |
| ENOX1 | CNOX | FLJ10094 | PIG38 | bA64J21.1 | cCNOX | ecto-NOX disulfide-thiol exchanger 1 | Two-hybrid | BioGRID | 16189514 |
| EPN1 | - | epsin 1 | - | HPRD | 10791968 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | - | HPRD,BioGRID | 14521715 |
| FAM101B | MGC45871 | family with sequence similarity 101, member B | Two-hybrid | BioGRID | 16169070 |
| FCHO1 | KIAA0290 | FCH domain only 1 | Two-hybrid | BioGRID | 16169070 |
| FHL2 | AAG11 | DRAL | SLIM3 | four and a half LIM domains 2 | - | HPRD,BioGRID | 12145280 |
| GATA1 | ERYF1 | GF-1 | GF1 | NFE1 | GATA binding protein 1 (globin transcription factor 1) | - | HPRD,BioGRID | 12242665 |
| GATA2 | MGC2306 | NFE1B | GATA binding protein 2 | - | HPRD,BioGRID | 11964310 |
| GCSH | GCE | NKH | glycine cleavage system protein H (aminomethyl carrier) | Two-hybrid | BioGRID | 16169070 |
| GSTM4 | GSTM4-4 | GTM4 | MGC131945 | MGC9247 | glutathione S-transferase mu 4 | Two-hybrid | BioGRID | 16169070 |
| HBEGF | DTR | DTS | DTSF | HEGFL | heparin-binding EGF-like growth factor | - | HPRD,BioGRID | 14597771 |15219838 |
| HBXIP | MGC71071 | XIP | hepatitis B virus x interacting protein | Two-hybrid | BioGRID | 16169070 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | - | HPRD | 9627120 |15467736 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 9627120 |9765306 |15467736 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | HDAC1 interacts with PLZF. This interaction was modelled on a demonstrated interaction between HDAC1 and PLZF from unspecified species. | BIND | 9486654 |
| HDAC2 | RPD3 | YAF1 | histone deacetylase 2 | Reconstituted Complex | BioGRID | 15467736 |
| HDAC3 | HD3 | RPD3 | RPD3-2 | histone deacetylase 3 | Reconstituted Complex | BioGRID | 15467736 |
| HDAC4 | HA6116 | HD4 | HDAC-A | HDACA | KIAA0288 | histone deacetylase 4 | - | HPRD,BioGRID | 11929873 |15467736 |
| HDAC5 | FLJ90614 | HD5 | NY-CO-9 | histone deacetylase 5 | - | HPRD,BioGRID | 11929873 |15467736 |
| HDAC6 | FLJ16239 | HD6 | JM21 | histone deacetylase 6 | - | HPRD,BioGRID | 15467736 |
| HDAC7 | DKFZp586J0917 | FLJ99588 | HD7A | HDAC7A | histone deacetylase 7 | Reconstituted Complex | BioGRID | 11929873 |
| HDAC9 | DKFZp779K1053 | HD7 | HDAC | HDAC7 | HDAC7B | HDAC9B | HDAC9FL | HDRP | KIAA0744 | MITR | histone deacetylase 9 | - | HPRD | 12590135 |
| HEATR2 | FLJ20397 | FLJ25564 | FLJ31671 | FLJ39381 | HEAT repeat containing 2 | Two-hybrid | BioGRID | 16169070 |
| KRTAP4-12 | KAP4.12 | KRTAP4.12 | keratin associated protein 4-12 | Two-hybrid | BioGRID | 16189514 |
| LDOC1 | BCUR1 | Mar7 | Mart7 | leucine zipper, down-regulated in cancer 1 | Two-hybrid | BioGRID | 16189514 |
| LMTK3 | KIAA1883 | LMR3 | TYKLM3 | lemur tyrosine kinase 3 | Two-hybrid | BioGRID | 16169070 |
| LSM2 | C6orf28 | G7b | YBL026W | snRNP | LSM2 homolog, U6 small nuclear RNA associated (S. cerevisiae) | Two-hybrid | BioGRID | 16169070 |
| LYAR | FLJ20425 | ZLYAR | Ly1 antibody reactive homolog (mouse) | Two-hybrid | BioGRID | 16169070 |
| MAGEA11 | MAGE-11 | MAGE11 | MAGEA-11 | MGC10511 | melanoma antigen family A, 11 | Two-hybrid | BioGRID | 16189514 |
| MAP3K3 | MAPKKK3 | MEKK3 | mitogen-activated protein kinase kinase kinase 3 | Two-hybrid | BioGRID | 16169070 |
| NCOR1 | KIAA1047 | MGC104216 | N-CoR | TRAC1 | hCIT529I10 | hN-CoR | nuclear receptor co-repressor 1 | - | HPRD,BioGRID | 9531570 |
| NCOR2 | CTG26 | SMRT | SMRTE | SMRTE-tau | TNRC14 | TRAC-1 | TRAC1 | nuclear receptor co-repressor 2 | - | HPRD | 9256429 |14982881 |
| NCOR2 | CTG26 | SMRT | SMRTE | SMRTE-tau | TNRC14 | TRAC-1 | TRAC1 | nuclear receptor co-repressor 2 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 9256429 |9765306 |14982881 |
| NCOR2 | CTG26 | SMRT | SMRTE | SMRTE-tau | TNRC14 | TRAC-1 | TRAC1 | nuclear receptor co-repressor 2 | SMRT interacts with PLZF. This interaction was modelled on a demonstrated interaction between SMRT and PLZF from unspecified species. | BIND | 9486654 |
| NR3C1 | GCCR | GCR | GR | GRL | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | - | HPRD,BioGRID | 14521715 |
| PAFAH1B3 | - | platelet-activating factor acetylhydrolase, isoform Ib, gamma subunit 29kDa | Two-hybrid | BioGRID | 16169070 |
| PGAM5 | BXLBv68 | MGC5352 | phosphoglycerate mutase family member 5 | Two-hybrid | BioGRID | 16169070 |
| PLSCR1 | MMTRA1B | phospholipid scramblase 1 | Two-hybrid | BioGRID | 16189514 |
| PMAIP1 | APR | NOXA | phorbol-12-myristate-13-acetate-induced protein 1 | Two-hybrid | BioGRID | 16169070 |
| PML | MYL | PP8675 | RNF71 | TRIM19 | promyelocytic leukemia | - | HPRD,BioGRID | 9294197 |11175338 |
| PSMD11 | MGC3844 | Rpn6 | S9 | p44.5 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 | Two-hybrid | BioGRID | 16169070 |
| PSMD2 | MGC14274 | P97 | Rpn1 | S2 | TRAP2 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 | Two-hybrid | BioGRID | 16169070 |
| QTRT1 | FP3235 | TGT | queuine tRNA-ribosyltransferase 1 | Two-hybrid | BioGRID | 16169070 |
| RAB27A | GS2 | HsT18676 | MGC117246 | RAB27 | RAM | RAB27A, member RAS oncogene family | Two-hybrid | BioGRID | 16169070 |
| RARA | NR1B1 | RAR | retinoic acid receptor, alpha | - | HPRD,BioGRID | 14521715 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | pRB interacts with PLZF. | BIND | 15949438 |
| RUNX1T1 | AML1T1 | CBFA2T1 | CDR | ETO | MGC2796 | MTG8 | MTG8b | ZMYND2 | runt-related transcription factor 1; translocated to, 1 (cyclin D-related) | - | HPRD,BioGRID | 10688654 |11090081 |
| RXRA | FLJ00280 | FLJ00318 | FLJ16020 | FLJ16733 | MGC102720 | NR2B1 | retinoid X receptor, alpha | - | HPRD | 8622986 |
| SIN3A | DKFZp434K2235 | FLJ90319 | KIAA0700 | SIN3 homolog A, transcription regulator (yeast) | - | HPRD | 9627120 |11719366 |
| SIN3A | DKFZp434K2235 | FLJ90319 | KIAA0700 | SIN3 homolog A, transcription regulator (yeast) | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 9627120 |9765306 |11719366 |
| SIN3A | DKFZp434K2235 | FLJ90319 | KIAA0700 | SIN3 homolog A, transcription regulator (yeast) | Sin3A interacts with PLZF. This interaction was modelled on a demonstrated interaction between mSin3A and PLZF from unspecified species. | BIND | 9486654 |
| SIN3B | KIAA0700 | SIN3 homolog B, transcription regulator (yeast) | - | HPRD,BioGRID | 9627120 |
| SP1 | - | Sp1 transcription factor | - | HPRD | 12004059 |
| SPOP | TEF2 | speckle-type POZ protein | Two-hybrid | BioGRID | 16189514 |
| TERF1 | FLJ41416 | PIN2 | TRBF1 | TRF | TRF1 | hTRF1-AS | t-TRF1 | telomeric repeat binding factor (NIMA-interacting) 1 | Two-hybrid | BioGRID | 16169070 |
| THNSL2 | FLJ10916 | THS2 | TSH2 | threonine synthase-like 2 (S. cerevisiae) | Two-hybrid | BioGRID | 16169070 |
| TOLLIP | FLJ33531 | IL-1RAcPIP | toll interacting protein | Two-hybrid | BioGRID | 16169070 |
| TRAF2 | MGC:45012 | TRAP | TRAP3 | TNF receptor-associated factor 2 | Two-hybrid | BioGRID | 16189514 |
| TRIM27 | RFP | RNF76 | tripartite motif-containing 27 | Two-hybrid | BioGRID | 16189514 |
| TXNIP | EST01027 | HHCPA78 | THIF | VDUP1 | thioredoxin interacting protein | - | HPRD,BioGRID | 12821938 |
| VDR | NR1I1 | vitamin D (1,25- dihydroxyvitamin D3) receptor | - | HPRD,BioGRID | 11719366 |12460926 |14521715 |
| VTA1 | C6orf55 | DRG-1 | DRG1 | FLJ27228 | HSPC228 | LIP5 | My012 | SBP1 | Vps20-associated 1 homolog (S. cerevisiae) | Two-hybrid | BioGRID | 16189514 |
| WDR33 | FLJ11294 | WDC146 | WD repeat domain 33 | Two-hybrid | BioGRID | 16169070 |
| ZBTB16 | PLZF | ZNF145 | zinc finger and BTB domain containing 16 | - | HPRD,BioGRID | 8622986 |
| ZBTB32 | FAXF | FAZF | Rog | TZFP | ZNF538 | zinc finger and BTB domain containing 32 | - | HPRD,BioGRID | 10572087 |
| ZNF24 | KOX17 | RSG-A | ZNF191 | ZSCAN3 | Zfp191 | zinc finger protein 24 | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG ACUTE MYELOID LEUKEMIA | 60 | 47 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EGFR SMRTE PATHWAY | 11 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS I MHC MEDIATED ANTIGEN PROCESSING PRESENTATION | 251 | 156 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | 212 | 129 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST DUCTAL CARCINOMA VS LOBULAR NORMAL DN | 69 | 43 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL DN | 455 | 304 | All SZGR 2.0 genes in this pathway |
| DOANE RESPONSE TO ANDROGEN UP | 184 | 125 | All SZGR 2.0 genes in this pathway |
| GRABARCZYK BCL11B TARGETS UP | 81 | 40 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC DN | 187 | 115 | All SZGR 2.0 genes in this pathway |
| HAHTOLA SEZARY SYNDROM DN | 42 | 26 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES DN | 16 | 10 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| CHEBOTAEV GR TARGETS UP | 77 | 62 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION DN | 234 | 147 | All SZGR 2.0 genes in this pathway |
| MARTIN INTERACT WITH HDAC | 44 | 31 | All SZGR 2.0 genes in this pathway |
| MYLLYKANGAS AMPLIFICATION HOT SPOT 23 | 24 | 13 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS BASAL | 330 | 217 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER UP | 227 | 137 | All SZGR 2.0 genes in this pathway |
| GOUYER TATI TARGETS UP | 10 | 7 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 UP | 256 | 159 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 1 UP | 140 | 85 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 3 UP | 17 | 11 | All SZGR 2.0 genes in this pathway |
| CERVERA SDHB TARGETS 1 DN | 38 | 22 | All SZGR 2.0 genes in this pathway |
| LE EGR2 TARGETS DN | 108 | 84 | All SZGR 2.0 genes in this pathway |
| FERRANDO LYL1 NEIGHBORS | 15 | 12 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS UP | 126 | 84 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS DN | 366 | 238 | All SZGR 2.0 genes in this pathway |
| WANG IMMORTALIZED BY HOXA9 AND MEIS1 DN | 24 | 15 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| YAGI AML FAB MARKERS | 191 | 131 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| CHEN LVAD SUPPORT OF FAILING HEART UP | 103 | 69 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 4 | 48 | 32 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION NEOCORTEX DN | 88 | 58 | All SZGR 2.0 genes in this pathway |
| VARELA ZMPSTE24 TARGETS UP | 40 | 30 | All SZGR 2.0 genes in this pathway |
| SARTIPY NORMAL AT INSULIN RESISTANCE DN | 21 | 11 | All SZGR 2.0 genes in this pathway |
| SARRIO EPITHELIAL MESENCHYMAL TRANSITION DN | 154 | 101 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL A UP | 84 | 52 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER POOR SURVIVAL UP | 31 | 22 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| MATZUK SPERMATOGONIA | 24 | 17 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C D DN | 252 | 155 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A4 | 196 | 124 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 HCP WITH H3K27ME3 | 435 | 318 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA PCA1 UP | 101 | 66 | All SZGR 2.0 genes in this pathway |
| HAHTOLA CTCL CUTANEOUS | 26 | 19 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 8 | 49 | 36 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |