Gene Page: BTG2
Summary ?
| GeneID | 7832 |
| Symbol | BTG2 |
| Synonyms | PC3|TIS21 |
| Description | BTG family member 2 |
| Reference | MIM:601597|HGNC:HGNC:1131|Ensembl:ENSG00000159388|HPRD:03357|Vega:OTTHUMG00000035834 |
| Gene type | protein-coding |
| Map location | 1q32 |
| Sherlock p-value | 0.484 |
| Fetal beta | -0.259 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs697455 | chr1 | 202512075 | BTG2 | 7832 | 0.18 | cis | ||
| rs12059674 | chr1 | 57630277 | BTG2 | 7832 | 0 | trans | ||
| rs17100274 | chr1 | 77774412 | BTG2 | 7832 | 0.13 | trans | ||
| rs1796829 | chr1 | 239661818 | BTG2 | 7832 | 0.15 | trans | ||
| rs12034521 | chr1 | 239717679 | BTG2 | 7832 | 0 | trans | ||
| snp_a-1990354 | 0 | BTG2 | 7832 | 2.022E-4 | trans | |||
| rs17038084 | chr2 | 85192280 | BTG2 | 7832 | 0.02 | trans | ||
| rs17498115 | chr2 | 85249294 | BTG2 | 7832 | 0.18 | trans | ||
| rs6707117 | chr2 | 136855504 | BTG2 | 7832 | 0 | trans | ||
| snp_a-2198363 | 0 | BTG2 | 7832 | 0.04 | trans | |||
| rs2875492 | chr3 | 86006209 | BTG2 | 7832 | 0.01 | trans | ||
| rs9288980 | chr3 | 112964991 | BTG2 | 7832 | 0.08 | trans | ||
| rs3995908 | chr3 | 112998529 | BTG2 | 7832 | 0.08 | trans | ||
| rs7652801 | chr3 | 113008605 | BTG2 | 7832 | 0.08 | trans | ||
| rs4631023 | chr4 | 8101794 | BTG2 | 7832 | 0.2 | trans | ||
| rs10040715 | chr5 | 57594207 | BTG2 | 7832 | 0 | trans | ||
| rs16894149 | chr5 | 61013724 | BTG2 | 7832 | 0 | trans | ||
| snp_a-4217300 | 0 | BTG2 | 7832 | 3.675E-4 | trans | |||
| rs2141482 | chr5 | 136760477 | BTG2 | 7832 | 0.14 | trans | ||
| rs2189803 | chr5 | 137684661 | BTG2 | 7832 | 8.295E-8 | trans | ||
| rs2237175 | chr6 | 16573534 | BTG2 | 7832 | 0.14 | trans | ||
| rs10945911 | chr6 | 164047509 | BTG2 | 7832 | 0.03 | trans | ||
| rs1419793 | chr7 | 34697630 | BTG2 | 7832 | 0.07 | trans | ||
| rs2199402 | chr8 | 9201002 | BTG2 | 7832 | 0.1 | trans | ||
| rs1381350 | chr8 | 9217194 | BTG2 | 7832 | 0.04 | trans | ||
| rs7841407 | chr8 | 9243427 | BTG2 | 7832 | 0.11 | trans | ||
| rs1733950 | chr8 | 90062516 | BTG2 | 7832 | 0.03 | trans | ||
| rs318307 | chr8 | 90185459 | BTG2 | 7832 | 0.06 | trans | ||
| rs9298651 | chr9 | 10076275 | BTG2 | 7832 | 0.02 | trans | ||
| rs16931013 | chr9 | 10076859 | BTG2 | 7832 | 0.02 | trans | ||
| rs16931018 | chr9 | 10077048 | BTG2 | 7832 | 0.02 | trans | ||
| rs6537944 | chr9 | 137312827 | BTG2 | 7832 | 0.09 | trans | ||
| rs4149623 | chr12 | 6450577 | BTG2 | 7832 | 0.1 | trans | ||
| rs2856339 | chr12 | 11906818 | BTG2 | 7832 | 0.09 | trans | ||
| rs8490 | chr12 | 51453904 | BTG2 | 7832 | 0.08 | trans | ||
| rs4905035 | chr14 | 93489262 | BTG2 | 7832 | 5.32E-4 | trans | ||
| rs941539 | chr14 | 93561214 | BTG2 | 7832 | 0.04 | trans | ||
| rs17128893 | chr14 | 93615606 | BTG2 | 7832 | 0.2 | trans | ||
| rs1885073 | chr14 | 104357181 | BTG2 | 7832 | 6.448E-4 | trans | ||
| rs8047151 | chr16 | 17209696 | BTG2 | 7832 | 0.1 | trans | ||
| rs5768165 | chr22 | 48318197 | BTG2 | 7832 | 0.02 | trans | ||
| rs5768168 | chr22 | 48319334 | BTG2 | 7832 | 0.02 | trans | ||
| rs5768173 | chr22 | 48321475 | BTG2 | 7832 | 0.02 | trans |
Section II. Transcriptome annotation
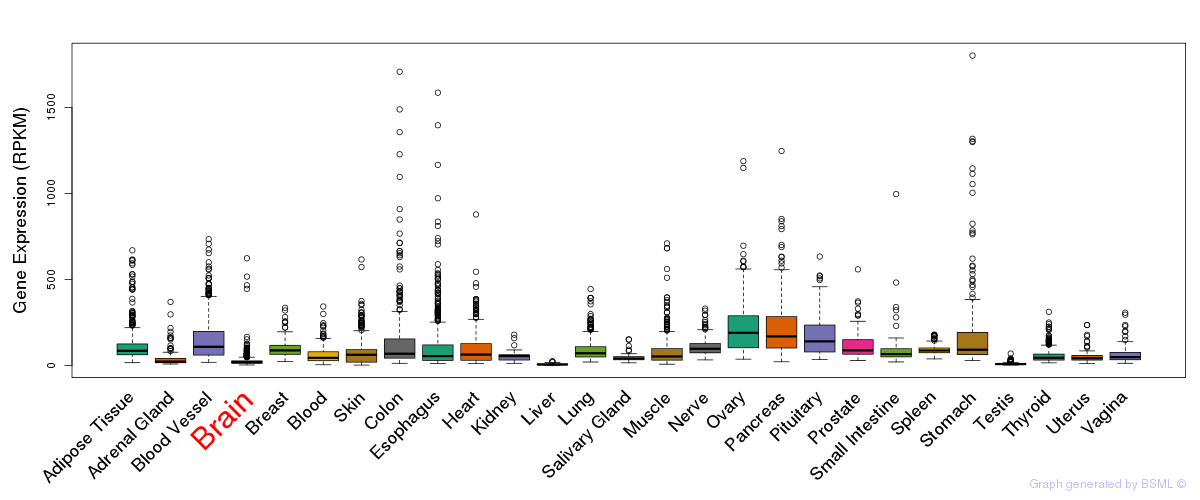
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID P53 DOWNSTREAM PATHWAY | 137 | 94 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP | 579 | 346 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE UP | 485 | 293 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL UP | 276 | 187 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA DN | 116 | 79 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL UP | 233 | 161 | All SZGR 2.0 genes in this pathway |
| LOPEZ MESOTELIOMA SURVIVAL TIME DN | 7 | 7 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION MONOCYTE DN | 54 | 38 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA UP | 544 | 308 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LPS UP | 431 | 237 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES CD4 UP | 64 | 46 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 48HR UP | 128 | 95 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 96HR UP | 117 | 84 | All SZGR 2.0 genes in this pathway |
| GESERICK TERT TARGETS DN | 21 | 16 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN UP | 612 | 367 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS F UP | 185 | 119 | All SZGR 2.0 genes in this pathway |
| KERLEY RESPONSE TO CISPLATIN UP | 44 | 30 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 2 | 127 | 92 | All SZGR 2.0 genes in this pathway |
| RASHI NFKB1 TARGETS | 19 | 18 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS LUMINAL | 326 | 213 | All SZGR 2.0 genes in this pathway |
| LUI THYROID CANCER CLUSTER 1 | 51 | 33 | All SZGR 2.0 genes in this pathway |
| DIRMEIER LMP1 RESPONSE EARLY | 66 | 48 | All SZGR 2.0 genes in this pathway |
| AMUNDSON DNA DAMAGE RESPONSE TP53 | 16 | 9 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 UP | 309 | 199 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER UP | 227 | 137 | All SZGR 2.0 genes in this pathway |
| BORLAK LIVER CANCER EGF UP | 57 | 41 | All SZGR 2.0 genes in this pathway |
| TSAI RESPONSE TO IONIZING RADIATION | 149 | 101 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM2 | 153 | 102 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| KHETCHOUMIAN TRIM24 TARGETS UP | 47 | 38 | All SZGR 2.0 genes in this pathway |
| FALVELLA SMOKERS WITH LUNG CANCER | 80 | 52 | All SZGR 2.0 genes in this pathway |
| AMIT DELAYED EARLY GENES | 18 | 15 | All SZGR 2.0 genes in this pathway |
| PASQUALUCCI LYMPHOMA BY GC STAGE UP | 283 | 177 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 20 HELA | 11 | 8 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 40 HELA | 42 | 29 | All SZGR 2.0 genes in this pathway |
| WANG PROSTATE CANCER ANDROGEN INDEPENDENT | 66 | 37 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO IKK INHIBITOR AND TNF UP | 223 | 140 | All SZGR 2.0 genes in this pathway |
| MORI PRE BI LYMPHOCYTE DN | 77 | 49 | All SZGR 2.0 genes in this pathway |
| MORI SMALL PRE BII LYMPHOCYTE UP | 86 | 57 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 1 UP | 140 | 85 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS UP | 266 | 171 | All SZGR 2.0 genes in this pathway |
| SHEPARD CRUSH AND BURN MUTANT UP | 197 | 110 | All SZGR 2.0 genes in this pathway |
| PENG LEUCINE DEPRIVATION UP | 142 | 93 | All SZGR 2.0 genes in this pathway |
| KANNAN TP53 TARGETS UP | 58 | 40 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC TGFA UP | 61 | 40 | All SZGR 2.0 genes in this pathway |
| KLEIN PRIMARY EFFUSION LYMPHOMA DN | 58 | 42 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| PEART HDAC PROLIFERATION CLUSTER UP | 57 | 35 | All SZGR 2.0 genes in this pathway |
| JECHLINGER EPITHELIAL TO MESENCHYMAL TRANSITION DN | 66 | 47 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER METASTASIS UP | 56 | 31 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE UP | 203 | 130 | All SZGR 2.0 genes in this pathway |
| HESS TARGETS OF HOXA9 AND MEIS1 DN | 77 | 48 | All SZGR 2.0 genes in this pathway |
| LENAOUR DENDRITIC CELL MATURATION DN | 128 | 90 | All SZGR 2.0 genes in this pathway |
| HADDAD B LYMPHOCYTE PROGENITOR | 293 | 193 | All SZGR 2.0 genes in this pathway |
| GERY CEBP TARGETS | 126 | 90 | All SZGR 2.0 genes in this pathway |
| BASSO CD40 SIGNALING DN | 68 | 44 | All SZGR 2.0 genes in this pathway |
| PETROVA ENDOTHELIUM LYMPHATIC VS BLOOD DN | 162 | 102 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS UP | 214 | 133 | All SZGR 2.0 genes in this pathway |
| TAVOR CEBPA TARGETS UP | 48 | 36 | All SZGR 2.0 genes in this pathway |
| BRACHAT RESPONSE TO CISPLATIN | 22 | 11 | All SZGR 2.0 genes in this pathway |
| GENTILE UV LOW DOSE UP | 27 | 19 | All SZGR 2.0 genes in this pathway |
| MCDOWELL ACUTE LUNG INJURY UP | 45 | 29 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO CANTHARIDIN UP | 19 | 12 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| CHEN LVAD SUPPORT OF FAILING HEART UP | 103 | 69 | All SZGR 2.0 genes in this pathway |
| BRACHAT RESPONSE TO METHOTREXATE UP | 26 | 16 | All SZGR 2.0 genes in this pathway |
| EHRLICH ICF SYNDROM DN | 15 | 13 | All SZGR 2.0 genes in this pathway |
| BRACHAT RESPONSE TO CAMPTOTHECIN UP | 28 | 18 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS PEAK AT 2HR | 51 | 36 | All SZGR 2.0 genes in this pathway |
| VARELA ZMPSTE24 TARGETS UP | 40 | 30 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER T4 | 94 | 69 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL A UP | 84 | 52 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| KUMAMOTO RESPONSE TO NUTLIN 3A UP | 9 | 5 | All SZGR 2.0 genes in this pathway |
| BONCI TARGETS OF MIR15A AND MIR16 1 | 91 | 75 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS DN | 210 | 128 | All SZGR 2.0 genes in this pathway |
| CHAUHAN RESPONSE TO METHOXYESTRADIOL DN | 102 | 65 | All SZGR 2.0 genes in this pathway |
| ICHIBA GRAFT VERSUS HOST DISEASE 35D UP | 131 | 79 | All SZGR 2.0 genes in this pathway |
| LEE DIFFERENTIATING T LYMPHOCYTE | 200 | 115 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR UP | 221 | 150 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR UP | 293 | 203 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS UP | 108 | 78 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A12 | 317 | 177 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING UP | 93 | 62 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS DN | 306 | 191 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR DN | 505 | 328 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS UP | 217 | 131 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS GROUP2 | 60 | 38 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS NOT VIA AKT1 DN | 88 | 61 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B | 549 | 316 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE MIDDLE | 98 | 56 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS DN | 315 | 215 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| JUBAN TARGETS OF SPI1 AND FLI1 UP | 115 | 73 | All SZGR 2.0 genes in this pathway |
| SMIRNOV RESPONSE TO IR 6HR UP | 166 | 97 | All SZGR 2.0 genes in this pathway |
| WARTERS RESPONSE TO IR SKIN | 83 | 44 | All SZGR 2.0 genes in this pathway |
| WARTERS IR RESPONSE 5GY | 47 | 23 | All SZGR 2.0 genes in this pathway |
| ZWANG DOWN BY 2ND EGF PULSE | 293 | 119 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |