Gene Page: HIST2H2BE
Summary ?
| GeneID | 8349 |
| Symbol | HIST2H2BE |
| Synonyms | GL105|H2B|H2B.1|H2BFQ|H2BGL105|H2BQ |
| Description | histone cluster 2, H2be |
| Reference | MIM:601831|HGNC:HGNC:4760|Ensembl:ENSG00000184678|HPRD:03494|Vega:OTTHUMG00000012095 |
| Gene type | protein-coding |
| Map location | 1q21.2 |
| Sherlock p-value | 0.54 |
| Fetal beta | 1.092 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0235 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00814 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg07015494 | 1 | 149857049 | HIST2H2BE;HIST2H2AC | 5.833E-4 | 0.365 | 0.049 | DMG:Wockner_2014 |
| cg07015494 | 1 | 149857049 | HIST2H2BE | 7.18E-8 | -0.017 | 1.72E-5 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs12096089 | chr1 | 112349065 | HIST2H2BE | 8349 | 0.14 | trans | ||
| rs6829546 | chr4 | 12587664 | HIST2H2BE | 8349 | 0.13 | trans | ||
| rs6448932 | chr4 | 12595798 | HIST2H2BE | 8349 | 0.15 | trans | ||
| rs4379216 | chr5 | 177511275 | HIST2H2BE | 8349 | 0.04 | trans | ||
| rs1917357 | chr7 | 124268065 | HIST2H2BE | 8349 | 0.16 | trans | ||
| rs11656351 | chr17 | 9489574 | HIST2H2BE | 8349 | 0.01 | trans | ||
| rs6637217 | chrX | 143443905 | HIST2H2BE | 8349 | 0.17 | trans |
Section II. Transcriptome annotation
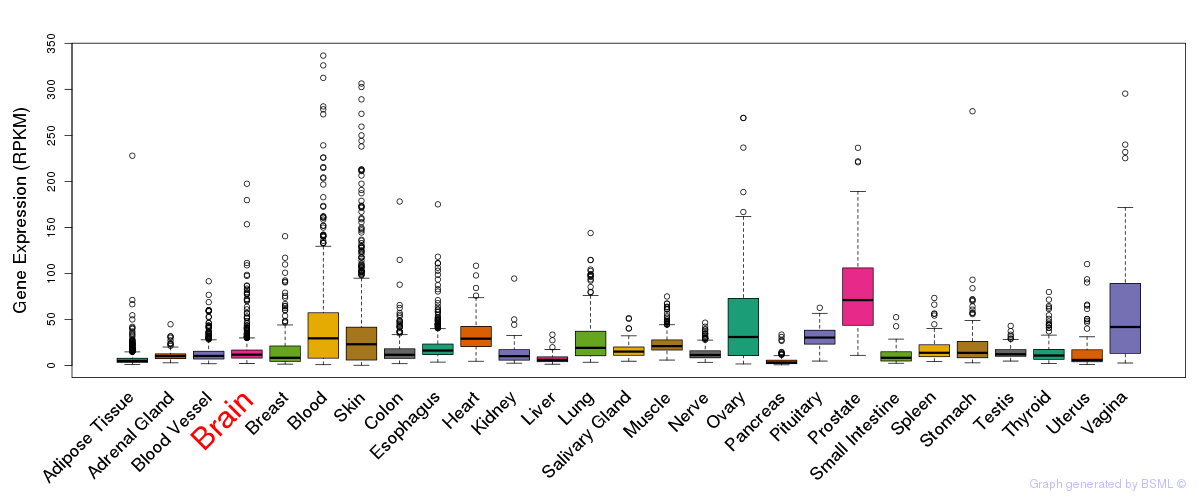
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CLIC1 | 0.74 | 0.65 |
| EFNA4 | 0.71 | 0.45 |
| CNN3 | 0.71 | 0.46 |
| VIM | 0.70 | 0.45 |
| TGIF1 | 0.69 | 0.42 |
| RFXANK | 0.68 | 0.60 |
| IQGAP2 | 0.68 | 0.22 |
| LRRC3B | 0.67 | 0.47 |
| FKBP10 | 0.67 | 0.48 |
| PDLIM3 | 0.67 | 0.50 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SYNJ2 | -0.32 | -0.35 |
| ITPR1 | -0.31 | -0.35 |
| TDRD6 | -0.31 | -0.40 |
| JAKMIP3 | -0.31 | -0.35 |
| ITSN2 | -0.31 | -0.39 |
| KIF16B | -0.31 | -0.34 |
| PIP5K2A | -0.30 | -0.34 |
| GPATCH4 | -0.30 | -0.40 |
| GREM1 | -0.29 | -0.33 |
| MYH7B | -0.29 | -0.26 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003677 | DNA binding | NAS | 1469070 | |
| GO:0005515 | protein binding | IPI | 17353931 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006334 | nucleosome assembly | NAS | 1469070 | |
| GO:0042742 | defense response to bacterium | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000786 | nucleosome | NAS | 1469070 | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005694 | chromosome | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BRD7 | BP75 | CELTIX1 | NAG4 | bromodomain containing 7 | - | HPRD | 12489984 |
| DNTTIP2 | ERBP | FCF2 | HSU15552 | LPTS-RP2 | MGC163494 | RP4-561L24.1 | TdIF2 | deoxynucleotidyltransferase, terminal, interacting protein 2 | - | HPRD | 12786946 |
| DYRK2 | FLJ21217 | FLJ21365 | dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 | - | HPRD | 9748265 |
| HIRA | DGCR1 | TUP1 | TUPLE1 | HIR histone cell cycle regulation defective homolog A (S. cerevisiae) | - | HPRD | 9710638 |
| HIRIP3 | - | HIRA interacting protein 3 | - | HPRD | 9710638 |
| HSPD1 | CPN60 | GROEL | HSP60 | HSP65 | HuCHA60 | SPG13 | heat shock 60kDa protein 1 (chaperonin) | - | HPRD,BioGRID | 9724719 |
| KPNA1 | IPOA5 | NPI-1 | RCH2 | SRP1 | karyopherin alpha 1 (importin alpha 5) | - | HPRD | 11824786 |
| LALBA | MGC138521 | MGC138523 | lactalbumin, alpha- | - | HPRD | 12888554 |
| NAP1L4 | MGC4565 | NAP2 | NAP2L | hNAP2 | nucleosome assembly protein 1-like 4 | - | HPRD | 9325046 |
| PARP3 | ADPRT3 | ADPRTL2 | ADPRTL3 | IRT1 | hPARP-3 | pADPRT-3 | poly (ADP-ribose) polymerase family, member 3 | Affinity Capture-MS | BioGRID | 17353931 |
| PTMA | MGC104802 | TMSA | prothymosin, alpha | - | HPRD | 11310559 |
| RCC1 | CHC1 | RCC1-I | regulator of chromosome condensation 1 | - | HPRD | 11375490 |
| TBL1X | EBI | SMAP55 | TBL1 | transducin (beta)-like 1X-linked | H2B interacts with TBL1. | BIND | 12628926 |
| TBL1X | EBI | SMAP55 | TBL1 | transducin (beta)-like 1X-linked | - | HPRD,BioGRID | 12628926 |
| TBL1XR1 | C21 | DC42 | FLJ12894 | IRA1 | TBLR1 | transducin (beta)-like 1 X-linked receptor 1 | H2B interacts with TBLR1. | BIND | 12628926 |
| TBL1XR1 | C21 | DC42 | FLJ12894 | IRA1 | TBLR1 | transducin (beta)-like 1 X-linked receptor 1 | Reconstituted Complex | BioGRID | 12628926 |
| TGM2 | G-ALPHA-h | GNAH | TG2 | TGC | transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase) | - | HPRD | 9973324 |
| TNPO1 | IPO2 | KPNB2 | MIP | MIP1 | TRN | transportin 1 | - | HPRD | 11824786 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG SYSTEMIC LUPUS ERYTHEMATOSUS | 140 | 100 | All SZGR 2.0 genes in this pathway |
| REACTOME MEIOSIS | 116 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I TRANSCRIPTION | 89 | 64 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTION | 210 | 127 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | 122 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME CHROMOSOME MAINTENANCE | 122 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | 64 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I PROMOTER OPENING | 62 | 49 | All SZGR 2.0 genes in this pathway |
| REACTOME MEIOTIC RECOMBINATION | 86 | 62 | All SZGR 2.0 genes in this pathway |
| REACTOME MEIOTIC SYNAPSIS | 73 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME AMYLOIDS | 83 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME PACKAGING OF TELOMERE ENDS | 48 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME TELOMERE MAINTENANCE | 75 | 57 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA DN | 349 | 157 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA UP | 177 | 110 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER DN | 406 | 230 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION DN | 329 | 219 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML QUIESCENT VS NORMAL QUIESCENT DN | 47 | 32 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML DIVIDING VS NORMAL QUIESCENT DN | 95 | 57 | All SZGR 2.0 genes in this pathway |
| GRAHAM NORMAL QUIESCENT VS NORMAL DIVIDING UP | 66 | 47 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES CD4 UP | 64 | 46 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES UP | 19 | 15 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| SANCHEZ MDM2 TARGETS | 15 | 10 | All SZGR 2.0 genes in this pathway |
| HERNANDEZ ABERRANT MITOSIS BY DOCETACEL 4NM UP | 23 | 15 | All SZGR 2.0 genes in this pathway |
| HERNANDEZ MITOTIC ARREST BY DOCETAXEL 1 UP | 36 | 18 | All SZGR 2.0 genes in this pathway |
| HERNANDEZ ABERRANT MITOSIS BY DOCETACEL 2NM UP | 81 | 57 | All SZGR 2.0 genes in this pathway |
| XU HGF SIGNALING NOT VIA AKT1 48HR DN | 20 | 12 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO PACLITAXEL VIA MAPK8 UP | 14 | 9 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| PASQUALUCCI LYMPHOMA BY GC STAGE DN | 165 | 104 | All SZGR 2.0 genes in this pathway |
| BENPORATH OCT4 TARGETS | 290 | 172 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| GNATENKO PLATELET SIGNATURE | 48 | 28 | All SZGR 2.0 genes in this pathway |
| FRASOR RESPONSE TO ESTRADIOL DN | 82 | 52 | All SZGR 2.0 genes in this pathway |
| REN BOUND BY E2F | 61 | 40 | All SZGR 2.0 genes in this pathway |
| VERNELL RETINOBLASTOMA PATHWAY DN | 22 | 13 | All SZGR 2.0 genes in this pathway |
| ALCALAY AML BY NPM1 LOCALIZATION UP | 140 | 83 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA UP | 64 | 46 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED UP | 183 | 111 | All SZGR 2.0 genes in this pathway |
| NOUZOVA TRETINOIN AND H4 ACETYLATION | 143 | 85 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION UP | 314 | 201 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE DN | 123 | 76 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK UP | 244 | 151 | All SZGR 2.0 genes in this pathway |
| ZHENG RESPONSE TO ARSENITE UP | 18 | 14 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE DN | 261 | 183 | All SZGR 2.0 genes in this pathway |
| GEORGANTAS HSC MARKERS | 71 | 47 | All SZGR 2.0 genes in this pathway |
| LIANG SILENCED BY METHYLATION UP | 32 | 18 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C3 | 20 | 15 | All SZGR 2.0 genes in this pathway |
| DAZARD UV RESPONSE CLUSTER G2 | 30 | 18 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS UP | 317 | 208 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| SARRIO EPITHELIAL MESENCHYMAL TRANSITION DN | 154 | 101 | All SZGR 2.0 genes in this pathway |
| CAMPS COLON CANCER COPY NUMBER DN | 74 | 34 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN | 270 | 181 | All SZGR 2.0 genes in this pathway |
| MOLENAAR TARGETS OF CCND1 AND CDK4 UP | 67 | 48 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| RAY TARGETS OF P210 BCR ABL FUSION UP | 18 | 13 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G3 UP | 188 | 121 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| KOBAYASHI EGFR SIGNALING 24HR UP | 101 | 65 | All SZGR 2.0 genes in this pathway |
| HAHTOLA CTCL CUTANEOUS | 26 | 19 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| DAZARD UV RESPONSE CLUSTER G24 | 28 | 19 | All SZGR 2.0 genes in this pathway |
| DASU IL6 SIGNALING UP | 59 | 44 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE M G1 | 148 | 95 | All SZGR 2.0 genes in this pathway |
| RAGHAVACHARI PLATELET SPECIFIC GENES | 70 | 46 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL DN | 428 | 246 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-23 | 1095 | 1101 | 1A | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.