Gene Page: SMARCA5
Summary ?
| GeneID | 8467 |
| Symbol | SMARCA5 |
| Synonyms | ISWI|SNF2H|WCRF135|hISWI|hSNF2H |
| Description | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| Reference | MIM:603375|HGNC:HGNC:11101|Ensembl:ENSG00000153147|HPRD:04538|Vega:OTTHUMG00000161474 |
| Gene type | protein-coding |
| Map location | 4q31.1-q31.2 |
| Pascal p-value | 0.005 |
| Sherlock p-value | 0.857 |
| Fetal beta | 0.897 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | Ascano FMRP targets Chromatin Remodeling Genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg16091845 | 4 | 144434637 | SMARCA5 | -0.022 | 0.67 | DMG:Nishioka_2013 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6599250 | chr3 | 38784028 | SMARCA5 | 8467 | 0.17 | trans | ||
| rs4491879 | chr3 | 189373908 | SMARCA5 | 8467 | 0.12 | trans | ||
| rs6046959 | chr20 | 20588433 | SMARCA5 | 8467 | 0.18 | trans |
Section II. Transcriptome annotation
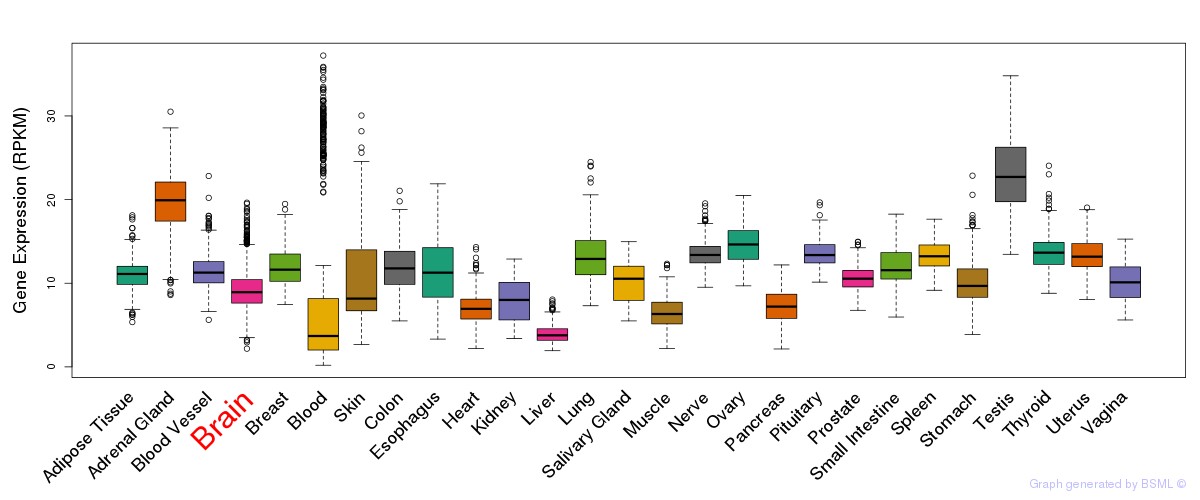
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MMADHC | 0.94 | 0.94 |
| DRG1 | 0.93 | 0.89 |
| PSMD14 | 0.93 | 0.90 |
| PPA1 | 0.93 | 0.90 |
| MRPL3 | 0.92 | 0.90 |
| TXNL1 | 0.92 | 0.90 |
| OLA1 | 0.92 | 0.90 |
| CCT4 | 0.92 | 0.94 |
| ATP5A1 | 0.92 | 0.90 |
| MRPS35 | 0.92 | 0.90 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.26 | -0.73 | -0.76 |
| AF347015.8 | -0.73 | -0.74 |
| AF347015.2 | -0.73 | -0.73 |
| MT-CYB | -0.73 | -0.73 |
| MT-CO2 | -0.73 | -0.71 |
| AF347015.33 | -0.72 | -0.74 |
| AF347015.15 | -0.72 | -0.74 |
| AF347015.31 | -0.70 | -0.69 |
| AF347015.27 | -0.68 | -0.70 |
| AF347015.18 | -0.65 | -0.74 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ALB | DKFZp779N1935 | PRO0883 | PRO0903 | PRO1341 | albumin | SNF2H interacts with the Albumin gene. | BIND | 15616580 |
| BAZ1A | ACF1 | DKFZp586E0518 | FLJ14383 | WALp1 | WCRF180 | hACF1 | bromodomain adjacent to zinc finger domain, 1A | - | HPRD,BioGRID | 10655480 |10880450 |11980720 |
| BAZ1A | ACF1 | DKFZp586E0518 | FLJ14383 | WALp1 | WCRF180 | hACF1 | bromodomain adjacent to zinc finger domain, 1A | SNF2H interacts with hACF1. | BIND | 15780937 |
| BAZ1B | WBSCR10 | WBSCR9 | WSTF | bromodomain adjacent to zinc finger domain, 1B | - | HPRD,BioGRID | 11980720 |
| BAZ2A | DKFZp781B109 | FLJ13768 | FLJ13780 | FLJ45876 | KIAA0314 | TIP5 | WALp3 | bromodomain adjacent to zinc finger domain, 2A | - | HPRD,BioGRID | 11532953 |
| CHD3 | Mi-2a | Mi2-ALPHA | ZFH | chromodomain helicase DNA binding protein 3 | Co-purification | BioGRID | 12198550 |
| CHRAC1 | CHARC1 | CHARC15 | CHRAC15 | YCL1 | chromatin accessibility complex 1 | - | HPRD | 12434153 |
| CHRAC1 | CHARC1 | CHARC15 | CHRAC15 | YCL1 | chromatin accessibility complex 1 | Affinity Capture-Western Co-purification | BioGRID | 10880450 |
| DNMT3B | ICF | M.HsaIIIB | DNA (cytosine-5-)-methyltransferase 3 beta | Co-purification | BioGRID | 15148359 |
| DNMT3B | ICF | M.HsaIIIB | DNA (cytosine-5-)-methyltransferase 3 beta | DNMT3B interacts with hSNF2H. | BIND | 15120635 |
| H3F3A | H3.3A | H3F3 | MGC87782 | MGC87783 | H3 histone, family 3A | H3F3A (Histone 3) interacts with SMARCA5 (SNF2H). | BIND | 15616580 |
| HDAC2 | RPD3 | YAF1 | histone deacetylase 2 | SMARCA5 (SNF2h) interacts with HDAC2. | BIND | 15775975 |
| HDAC2 | RPD3 | YAF1 | histone deacetylase 2 | Co-purification | BioGRID | 12198550 |
| HNF1A | HNF1 | LFB1 | MODY3 | TCF1 | HNF1 homeobox A | SNF2H interacts with the HNF-1 gene. | BIND | 15616580 |
| PHC3 | DKFZp313K1221 | EDR3 | FLJ12729 | FLJ12967 | HPH3 | MGC88144 | polyhomeotic homolog 3 (Drosophila) | - | HPRD | 12167701 |
| POLE3 | CHARAC17 | CHRAC17 | YBL1 | p17 | polymerase (DNA directed), epsilon 3 (p17 subunit) | Affinity Capture-Western Co-purification | BioGRID | 10880450 |
| RAD21 | FLJ25655 | FLJ40596 | HR21 | HRAD21 | KIAA0078 | MCD1 | NXP1 | SCC1 | hHR21 | RAD21 homolog (S. pombe) | - | HPRD,BioGRID | 12198550 |
| RBBP4 | NURF55 | RBAP48 | retinoblastoma binding protein 4 | Co-purification | BioGRID | 12198550 |
| SATB1 | - | SATB homeobox 1 | - | HPRD,BioGRID | 12374985 |
| SMC1A | CDLS2 | DKFZp686L19178 | DXS423E | KIAA0178 | MGC138332 | SB1.8 | SMC1 | SMC1L1 | SMC1alpha | SMCB | structural maintenance of chromosomes 1A | Co-purification | BioGRID | 12198550 |
| SMC3 | BAM | BMH | CDLS3 | CSPG6 | HCAP | SMC3L1 | structural maintenance of chromosomes 3 | Co-purification | BioGRID | 12198550 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME CHROMOSOME MAINTENANCE | 122 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | 64 | 43 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS UP | 332 | 228 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| BARIS THYROID CANCER DN | 59 | 45 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER SOX9 TARGETS IN PROSTATE DEVELOPMENT DN | 45 | 33 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION UP | 195 | 138 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS UP | 424 | 268 | All SZGR 2.0 genes in this pathway |
| MORI IMMATURE B LYMPHOCYTE DN | 90 | 55 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA CD1 AND CD2 UP | 89 | 51 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS DN | 371 | 218 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN DN | 271 | 175 | All SZGR 2.0 genes in this pathway |
| RIGGINS TAMOXIFEN RESISTANCE DN | 220 | 147 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS UP | 170 | 107 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS UP | 425 | 253 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR DN | 185 | 116 | All SZGR 2.0 genes in this pathway |
| WINNEPENNINCKX MELANOMA METASTASIS UP | 162 | 86 | All SZGR 2.0 genes in this pathway |
| ONGUSAHA BRCA1 TARGETS DN | 14 | 9 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |