Gene Page: KAT2B
Summary ?
| GeneID | 8850 |
| Symbol | KAT2B |
| Synonyms | CAF|P/CAF|PCAF |
| Description | lysine acetyltransferase 2B |
| Reference | MIM:602303|HGNC:HGNC:8638|Ensembl:ENSG00000114166|HPRD:06780|Vega:OTTHUMG00000130481 |
| Gene type | protein-coding |
| Map location | 3p24 |
| Pascal p-value | 0.19 |
| Fetal beta | -1.344 |
| Support | Chromatin Remodeling Genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
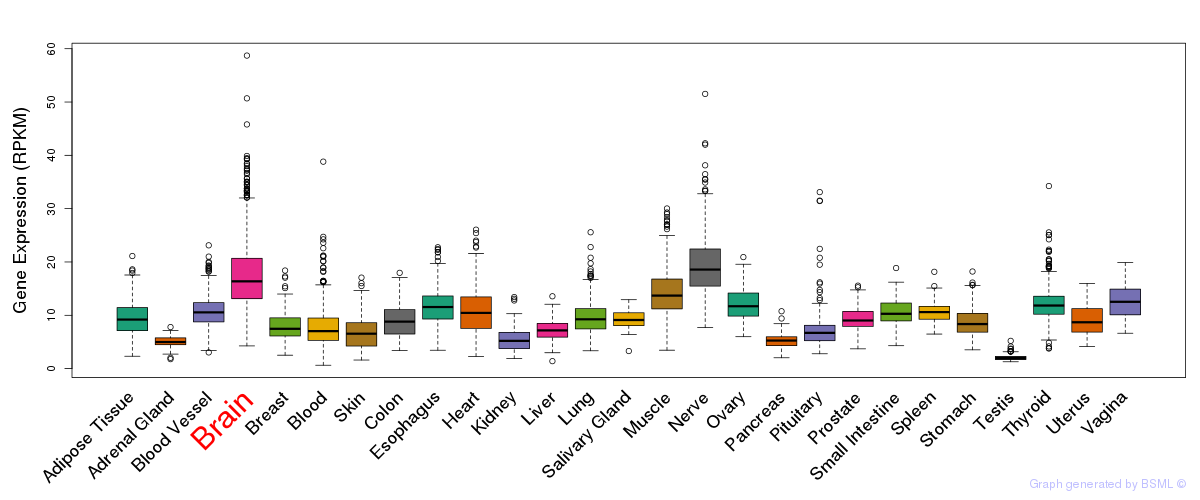
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003713 | transcription coactivator activity | IDA | 12435739 | |
| GO:0004402 | histone acetyltransferase activity | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0008080 | N-acetyltransferase activity | IEA | - | |
| GO:0042826 | histone deacetylase binding | IPI | 12887892 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0006338 | chromatin remodeling | NAS | 10365964 | |
| GO:0006473 | protein amino acid acetylation | TAS | 8684459 | |
| GO:0007049 | cell cycle | IEA | - | |
| GO:0007050 | cell cycle arrest | TAS | 8684459 | |
| GO:0008285 | negative regulation of cell proliferation | IDA | 8684459 | |
| GO:0008152 | metabolic process | IEA | - | |
| GO:0018076 | N-terminal peptidyl-lysine acetylation | IDA | 12435739 | |
| GO:0044419 | interspecies interaction between organisms | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | TAS | 10891508 | |
| GO:0005654 | nucleoplasm | EXP | 11250901 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ALB | DKFZp779N1935 | PRO0883 | PRO0903 | PRO1341 | albumin | PCAF interacts with the Albumin gene. | BIND | 15616580 |
| ARNTL | BMAL1 | BMAL1c | JAP3 | MGC47515 | MOP3 | PASD3 | TIC | bHLHe5 | aryl hydrocarbon receptor nuclear translocator-like | Reconstituted Complex | BioGRID | 14645221 |
| BRCA2 | BRCC2 | FACD | FAD | FAD1 | FANCB | FANCD | FANCD1 | breast cancer 2, early onset | - | HPRD,BioGRID | 9824164 |
| CCND1 | BCL1 | D11S287E | PRAD1 | U21B31 | cyclin D1 | - | HPRD,BioGRID | 10318892 |
| CDC25B | - | cell division cycle 25 homolog B (S. pombe) | - | HPRD,BioGRID | 11689696 |
| CDK8 | K35 | MGC126074 | MGC126075 | cyclin-dependent kinase 8 | Affinity Capture-Western | BioGRID | 9710619 |
| CLOCK | KAT13D | KIAA0334 | bHLHe8 | clock homolog (mouse) | Reconstituted Complex | BioGRID | 14645221 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | Affinity Capture-Western Co-fractionation Reconstituted Complex | BioGRID | 9710619 |11864601 |
| CTBP1 | BARS | MGC104684 | C-terminal binding protein 1 | CTBP1 interacts with PCAF | BIND | 15834423 |
| CTBP1 | BARS | MGC104684 | C-terminal binding protein 1 | Biochemical Activity | BioGRID | 9824164 |
| CUX1 | CASP | CDP | CDP/Cut | CDP1 | COY1 | CUTL1 | CUX | Clox | Cux/CDP | GOLIM6 | Nbla10317 | p100 | p110 | p200 | p75 | cut-like homeobox 1 | - | HPRD,BioGRID | 10852958 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | Biochemical Activity | BioGRID | 12419806 |
| EVI1 | AML1-EVI-1 | EVI-1 | MDS1-EVI1 | MGC163392 | PRDM3 | ecotropic viral integration site 1 | - | HPRD,BioGRID | 11568182 |
| H3F3A | H3.3A | H3F3 | MGC87782 | MGC87783 | H3 histone, family 3A | H3F3A (Histone 3) interacts with PCAF. | BIND | 15616580 |
| HIST4H4 | H4/p | MGC24116 | histone cluster 4, H4 | PCAF interacts with and acetylates H4. This interaction was modeled on a demonstrated interaction between PCAF and H4, both from unspecified species. | BIND | 15992539 |
| HNF1A | HNF1 | LFB1 | MODY3 | TCF1 | HNF1 homeobox A | - | HPRD,BioGRID | 10777539 |
| ING1 | p24ING1c | p33 | p33ING1 | p33ING1b | p47 | p47ING1a | inhibitor of growth family, member 1 | - | HPRD,BioGRID | 12015309 |
| IRF1 | IRF-1 | MAR | interferon regulatory factor 1 | - | HPRD,BioGRID | 10022868 |11304541 |
| IRF2 | DKFZp686F0244 | IRF-2 | interferon regulatory factor 2 | - | HPRD,BioGRID | 10022868 |11304541 |
| JDP2 | JUNDM2 | Jun dimerization protein 2 | - | HPRD | 12101239 |
| KLF13 | BTEB3 | FKLF2 | NSLP1 | RFLAT-1 | RFLAT1 | Kruppel-like factor 13 | - | HPRD,BioGRID | 11748222 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | - | HPRD,BioGRID | 12068014 |
| MED21 | SRB7 | SURB7 | mediator complex subunit 21 | Affinity Capture-Western | BioGRID | 9710619 |
| MYC | bHLHe39 | c-Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | Affinity Capture-Western Co-purification Reconstituted Complex | BioGRID | 12660246 |
| NCOA1 | F-SRC-1 | KAT13A | MGC129719 | MGC129720 | NCoA-1 | RIP160 | SRC-1 | SRC1 | bHLHe42 | nuclear receptor coactivator 1 | - | HPRD,BioGRID | 9296499 |
| NCOA3 | ACTR | AIB-1 | AIB1 | CAGH16 | CTG26 | KAT13B | MGC141848 | RAC3 | SRC3 | TNRC14 | TNRC16 | TRAM-1 | pCIP | nuclear receptor coactivator 3 | P/CAF interacts with TRAM-1. | BIND | 9346901 |
| NCOA3 | ACTR | AIB-1 | AIB1 | CAGH16 | CTG26 | KAT13B | MGC141848 | RAC3 | SRC3 | TNRC14 | TNRC16 | TRAM-1 | pCIP | nuclear receptor coactivator 3 | - | HPRD | 9445475 |
| NCOA4 | ARA70 | DKFZp762E1112 | ELE1 | PTC3 | RFG | nuclear receptor coactivator 4 | - | HPRD,BioGRID | 9892017 |
| NOTCH1 | TAN1 | hN1 | Notch homolog 1, translocation-associated (Drosophila) | - | HPRD | 10747963 |
| NOTCH3 | CADASIL | CASIL | Notch homolog 3 (Drosophila) | - | HPRD | 11404076 |
| NPAS2 | FLJ23138 | MGC71151 | MOP4 | PASD4 | bHLHe9 | neuronal PAS domain protein 2 | Reconstituted Complex | BioGRID | 14645221 |
| NR4A1 | GFRP1 | HMR | MGC9485 | N10 | NAK-1 | NGFIB | NP10 | NUR77 | TR3 | nuclear receptor subfamily 4, group A, member 1 | - | HPRD,BioGRID | 12082103 |
| NRIP1 | FLJ77253 | RIP140 | nuclear receptor interacting protein 1 | - | HPRD | 14581481 |
| ONECUT1 | HNF-6 | HNF6 | HNF6A | one cut homeobox 1 | - | HPRD,BioGRID | 10811635 |
| PGR | NR3C3 | PR | progesterone receptor | PRB interacts with PCAF. This interaction is modelled on a demonstrated interaction between human PRB and PCAF from an unspecified species. | BIND | 9223281 |
| POLR2A | MGC75453 | POLR2 | POLRA | RPB1 | RPBh1 | RPO2 | RPOL2 | RpIILS | hRPB220 | hsRPB1 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | Affinity Capture-Western | BioGRID | 9710619 |
| RARA | NR1B1 | RAR | retinoic acid receptor, alpha | Affinity Capture-Western | BioGRID | 14581481 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | pRb interacts with the acetyltransferase domain of P/CAF. | BIND | 15044952 |
| RBPJ | CBF1 | IGKJRB | IGKJRB1 | KBF2 | MGC61669 | RBP-J | RBPJK | RBPSUH | SUH | csl | recombination signal binding protein for immunoglobulin kappa J region | - | HPRD,BioGRID | 10747963 |
| SMARCB1 | BAF47 | INI1 | RDT | SNF5 | SNF5L1 | Sfh1p | Snr1 | hSNFS | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1 | Affinity Capture-Western | BioGRID | 9710619 |
| SRCAP | DOMO1 | EAF1 | FLJ44499 | KIAA0309 | SWR1 | Snf2-related CREBBP activator protein | - | HPRD | 11581372 |
| SUPT3H | SPT3 | SPT3L | suppressor of Ty 3 homolog (S. cerevisiae) | Affinity Capture-Western | BioGRID | 12660246 |
| TACC2 | AZU-1 | ECTACC | transforming, acidic coiled-coil containing protein 2 | TACC2 interacts with pCAF. | BIND | 14767476 |
| TAF5L | PAF65B | TAF5-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor, 65kDa | Affinity Capture-Western | BioGRID | 12660246 |
| TAF9 | AD-004 | AK6 | CGI-137 | CINAP | CIP | MGC1603 | MGC3647 | MGC5067 | MGC:1603 | MGC:3647 | MGC:5067 | TAF2G | TAFII31 | TAFII32 | TAFIID32 | TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa | Affinity Capture-Western | BioGRID | 12660246 |
| TCF3 | E2A | ITF1 | MGC129647 | MGC129648 | bHLHb21 | transcription factor 3 (E2A immunoglobulin enhancer binding factors E12/E47) | - | HPRD,BioGRID | 12435739 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | Biochemical Activity | BioGRID | 12068014 |
| TRRAP | FLJ10671 | PAF350/400 | PAF400 | STAF40 | TR-AP | Tra1 | transformation/transcription domain-associated protein | Affinity Capture-Western | BioGRID | 11509179 |12660246 |
| TTF1 | - | transcription termination factor, RNA polymerase I | - | HPRD,BioGRID | 11250901 |
| TWIST1 | ACS3 | BPES2 | BPES3 | SCS | TWIST | bHLHa38 | twist homolog 1 (Drosophila) | - | HPRD,BioGRID | 10025406 |
| YY1 | DELTA | INO80S | NF-E1 | UCRBP | YIN-YANG-1 | YY1 transcription factor | Biochemical Activity | BioGRID | 11486036 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NOTCH SIGNALING PATHWAY | 47 | 35 | All SZGR 2.0 genes in this pathway |
| BIOCARTA VDR PATHWAY | 12 | 7 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RARRXR PATHWAY | 15 | 9 | All SZGR 2.0 genes in this pathway |
| PID SMAD2 3NUCLEAR PATHWAY | 82 | 63 | All SZGR 2.0 genes in this pathway |
| PID HDAC CLASSIII PATHWAY | 25 | 20 | All SZGR 2.0 genes in this pathway |
| PID E2F PATHWAY | 74 | 48 | All SZGR 2.0 genes in this pathway |
| PID HDAC CLASSI PATHWAY | 66 | 50 | All SZGR 2.0 genes in this pathway |
| PID FOXO PATHWAY | 49 | 43 | All SZGR 2.0 genes in this pathway |
| PID AR TF PATHWAY | 53 | 38 | All SZGR 2.0 genes in this pathway |
| PID RETINOIC ACID PATHWAY | 30 | 23 | All SZGR 2.0 genes in this pathway |
| PID P53 REGULATION PATHWAY | 59 | 50 | All SZGR 2.0 genes in this pathway |
| REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | 29 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | 24 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME PRE NOTCH EXPRESSION AND PROCESSING | 44 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | 46 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH1 | 70 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME GENERIC TRANSCRIPTION PATHWAY | 352 | 181 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I TRANSCRIPTION | 89 | 64 | All SZGR 2.0 genes in this pathway |
| REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | 13 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTION | 210 | 127 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | 122 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH | 103 | 64 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I TRANSCRIPTION INITIATION | 25 | 14 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION MONOCYTE DN | 54 | 38 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP DN | 199 | 124 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| BERENJENO ROCK SIGNALING NOT VIA RHOA DN | 48 | 34 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| NAGY PCAF COMPONENTS HUMAN | 9 | 7 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS NEUROEPITHELIUM DN | 164 | 111 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER DN | 514 | 319 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER DN | 203 | 134 | All SZGR 2.0 genes in this pathway |
| PUJANA BREAST CANCER LIT INT NETWORK | 101 | 73 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 UP | 256 | 159 | All SZGR 2.0 genes in this pathway |
| MENSSEN MYC TARGETS | 53 | 35 | All SZGR 2.0 genes in this pathway |
| CHANG POU5F1 TARGETS UP | 15 | 10 | All SZGR 2.0 genes in this pathway |
| UEDA PERIFERAL CLOCK | 169 | 111 | All SZGR 2.0 genes in this pathway |
| HADDAD B LYMPHOCYTE PROGENITOR | 293 | 193 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP | 390 | 242 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS DN | 371 | 218 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| MULLIGAN NTF3 SIGNALING VIA INSR AND IGF1R DN | 13 | 6 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 6 | 189 | 112 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| STEARMAN LUNG CANCER EARLY VS LATE UP | 125 | 89 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN | 270 | 181 | All SZGR 2.0 genes in this pathway |
| HUANG FOXA2 TARGETS UP | 45 | 28 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN DN | 249 | 165 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| CHUNG BLISTER CYTOTOXICITY DN | 44 | 29 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G1 DN | 40 | 26 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA PR DN | 44 | 34 | All SZGR 2.0 genes in this pathway |
| NIELSEN GIST | 98 | 66 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE S | 162 | 86 | All SZGR 2.0 genes in this pathway |
| STAMBOLSKY RESPONSE TO VITAMIN D3 DN | 26 | 14 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 6HR DN | 91 | 56 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR DN | 505 | 328 | All SZGR 2.0 genes in this pathway |
| SUBTIL PROGESTIN TARGETS | 36 | 25 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND WITH H4K20ME1 MARK | 145 | 82 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM A | 182 | 108 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-142-3p | 1141 | 1148 | 1A,m8 | hsa-miR-142-3p | UGUAGUGUUUCCUACUUUAUGGA |
| miR-17-5p/20/93.mr/106/519.d | 120 | 126 | m8 | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-181 | 366 | 372 | m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-19 | 314 | 320 | 1A | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-200bc/429 | 1021 | 1027 | m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-205 | 420 | 426 | 1A | hsa-miR-205 | UCCUUCAUUCCACCGGAGUCUG |
| miR-25/32/92/363/367 | 1748 | 1755 | 1A,m8 | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| miR-27 | 1618 | 1624 | 1A | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-30-3p | 557 | 563 | 1A | hsa-miR-30a-3p | CUUUCAGUCGGAUGUUUGCAGC |
| hsa-miR-30e-3p | CUUUCAGUCGGAUGUUUACAGC | ||||
| miR-330 | 543 | 550 | 1A,m8 | hsa-miR-330brain | GCAAAGCACACGGCCUGCAGAGA |
| miR-495 | 1626 | 1633 | 1A,m8 | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
| miR-93.hd/291-3p/294/295/302/372/373/520 | 119 | 125 | m8 | hsa-miR-93brain | AAAGUGCUGUUCGUGCAGGUAG |
| hsa-miR-302a | UAAGUGCUUCCAUGUUUUGGUGA | ||||
| hsa-miR-302b | UAAGUGCUUCCAUGUUUUAGUAG | ||||
| hsa-miR-302c | UAAGUGCUUCCAUGUUUCAGUGG | ||||
| hsa-miR-302d | UAAGUGCUUCCAUGUUUGAGUGU | ||||
| hsa-miR-372 | AAAGUGCUGCGACAUUUGAGCGU | ||||
| hsa-miR-373 | GAAGUGCUUCGAUUUUGGGGUGU | ||||
| hsa-miR-520e | AAAGUGCUUCCUUUUUGAGGG | ||||
| hsa-miR-520a | AAAGUGCUUCCCUUUGGACUGU | ||||
| hsa-miR-520b | AAAGUGCUUCCUUUUAGAGGG | ||||
| hsa-miR-520c | AAAGUGCUUCCUUUUAGAGGGUU | ||||
| hsa-miR-520d | AAAGUGCUUCUCUUUGGUGGGUU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.