Gene Page: MAD1L1
Summary ?
| GeneID | 8379 |
| Symbol | MAD1L1 |
| Synonyms | MAD1|PIG9|TP53I9|TXBP181 |
| Description | MAD1 mitotic arrest deficient-like 1 (yeast) |
| Reference | MIM:602686|HGNC:HGNC:6762|Ensembl:ENSG00000002822|HPRD:04065|Vega:OTTHUMG00000151493 |
| Gene type | protein-coding |
| Map location | 7p22 |
| Pascal p-value | 5.049E-11 |
| Fetal beta | -0.594 |
| DMG | 3 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGC128 | Genome-wide Association Study | A multi-stage schizophrenia GWAS of up to 36,989 cases and 113,075 controls. Reported by the Schizophrenia Working Group of PGC. 128 independent associations spanning 108 loci | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| CV:Ripke_2013 | Genome-wide Association Study | Multi-stage GWAS, Sweden population and PGC2. 24 leading SNPs | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 4 |
| DMG:Montano_2016 | Genome-wide DNA methylation analysis | This dataset includes 172 replicated associations between CpGs with schizophrenia. | 4 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 4 |
| LK:YES | Genome-wide Association Study | This data set included 99 genes mapped to the 22 regions. The 24 leading SNPs were also included in CV:Ripke_2013 | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 CV:PGC128
CV:PGC128
| SNP ID | Chromosome | Position | Allele | P | Function | Gene | Up/Down Distance |
|---|
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg25323444 | 7 | 2111060 | MAD1L1 | 1.07E-6 | 0.011 | 0.016 | DMG:Montano_2016 |
| cg23997168 | 7 | 1984831 | MAD1L1 | 8.11E-5 | 0.323 | 0.026 | DMG:Wockner_2014 |
| cg04395498 | 7 | 2017437 | MAD1L1 | 2.208E-4 | 0.465 | 0.036 | DMG:Wockner_2014 |
| cg20935553 | 7 | 2272059 | MAD1L1 | 7.53E-9 | -0.015 | 3.72E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
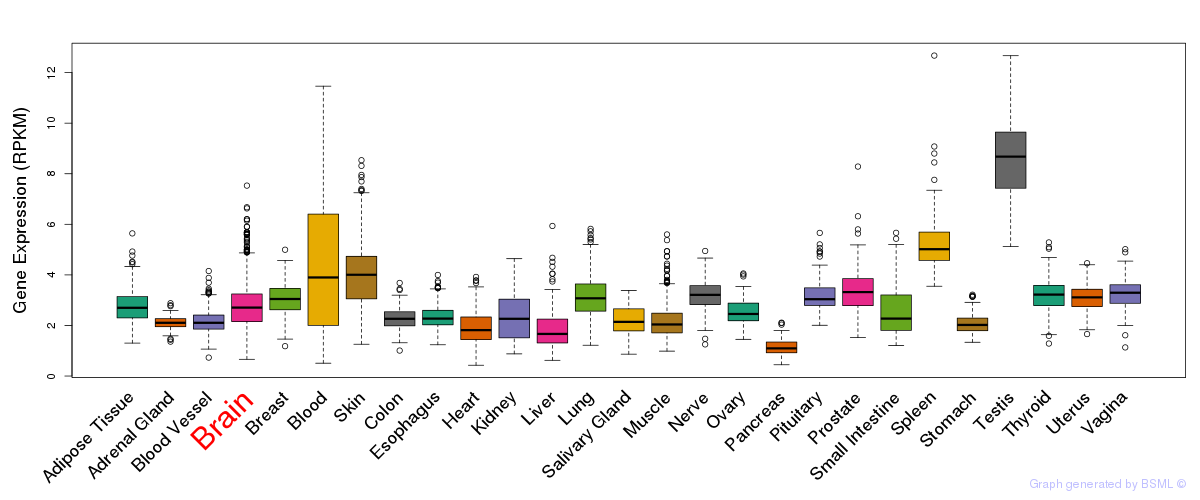
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RBM5 | 0.91 | 0.92 |
| ZC3H11A | 0.90 | 0.91 |
| EXOSC10 | 0.90 | 0.90 |
| SFRS14 | 0.89 | 0.91 |
| ZNF509 | 0.89 | 0.90 |
| WDR59 | 0.89 | 0.89 |
| INTS3 | 0.89 | 0.92 |
| HARS2 | 0.89 | 0.89 |
| ZNF862 | 0.88 | 0.89 |
| TCERG1 | 0.88 | 0.91 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.75 | -0.82 |
| MT-CO2 | -0.74 | -0.81 |
| AF347015.27 | -0.73 | -0.79 |
| AF347015.33 | -0.71 | -0.77 |
| IFI27 | -0.71 | -0.78 |
| MT-CYB | -0.70 | -0.77 |
| AF347015.21 | -0.70 | -0.84 |
| FXYD1 | -0.70 | -0.78 |
| AF347015.8 | -0.70 | -0.79 |
| HIGD1B | -0.70 | -0.80 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AMOTL2 | LCCP | angiomotin like 2 | Two-hybrid | BioGRID | 16189514 |
| FAM131C | C1orf117 | FLJ36766 | RP11-5P18.9 | family with sequence similarity 131, member C | Two-hybrid | BioGRID | 16189514 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | Affinity Capture-Western Co-purification | BioGRID | 15388328 |
| HDAC2 | RPD3 | YAF1 | histone deacetylase 2 | Affinity Capture-Western Co-purification | BioGRID | 15388328 |
| MAD1L1 | HsMAD1 | MAD1 | PIG9 | TP53I9 | TXBP181 | MAD1 mitotic arrest deficient-like 1 (yeast) | Two-hybrid | BioGRID | 16189514 |
| MAD2L1 | HSMAD2 | MAD2 | MAD2 mitotic arrest deficient-like 1 (yeast) | - | HPRD | 11804586 |
| MAD2L1 | HSMAD2 | MAD2 | MAD2 mitotic arrest deficient-like 1 (yeast) | MAD1L1 (Mad1) interacts with MAD2L1 (Mad2). | BIND | 15694304 |
| MAD2L1 | HSMAD2 | MAD2 | MAD2 mitotic arrest deficient-like 1 (yeast) | Reconstituted Complex Two-hybrid | BioGRID | 10660610 |11707408 |16189514 |
| MAD2L1 | HSMAD2 | MAD2 | MAD2 mitotic arrest deficient-like 1 (yeast) | TXBP181 interacts with HsMAD2. | BIND | 9546394 |
| NDC80 | HEC | HEC1 | KNTC2 | TID3 | hsNDC80 | NDC80 homolog, kinetochore complex component (S. cerevisiae) | - | HPRD | 12351790 |
| NONO | NMT55 | NRB54 | P54 | P54NRB | non-POU domain containing, octamer-binding | Two-hybrid | BioGRID | 16189514 |
| RP4-691N24.1 | FLJ11792 | KIAA0980 | NLP | dJ691N24.1 | ninein-like | Two-hybrid | BioGRID | 16189514 |
| SIN3A | DKFZp434K2235 | FLJ90319 | KIAA0700 | SIN3 homolog A, transcription regulator (yeast) | - | HPRD | 11106735 |
| SIN3B | KIAA0700 | SIN3 homolog B, transcription regulator (yeast) | - | HPRD | 11101889 |11370785 |
| TEX11 | TGC1 | TSGA3 | testis expressed 11 | Two-hybrid | BioGRID | 16189514 |
| TRIM29 | ATDC | FLJ36085 | tripartite motif-containing 29 | Two-hybrid | BioGRID | 16189514 |
| TUBGCP4 | 76P | FLJ14797 | GCP4 | tubulin, gamma complex associated protein 4 | Two-hybrid | BioGRID | 16189514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CELL CYCLE | 128 | 84 | All SZGR 2.0 genes in this pathway |
| PID CMYB PATHWAY | 84 | 61 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE CHECKPOINTS | 124 | 70 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC M M G1 PHASES | 172 | 98 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPLICATION | 192 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC PROMETAPHASE | 87 | 51 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL UP | 276 | 187 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS UP | 135 | 82 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| LANG MYB FAMILY TARGETS | 29 | 16 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 TTD DN | 84 | 63 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 480 HELA | 164 | 118 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 7P22 AMPLICON | 38 | 27 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS UP | 126 | 84 | All SZGR 2.0 genes in this pathway |
| FERRANDO T ALL WITH MLL ENL FUSION UP | 87 | 67 | All SZGR 2.0 genes in this pathway |
| HOFMANN CELL LYMPHOMA DN | 39 | 29 | All SZGR 2.0 genes in this pathway |
| XU RESPONSE TO TRETINOIN DN | 13 | 8 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 48HR DN | 161 | 105 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS | 212 | 121 | All SZGR 2.0 genes in this pathway |
| CHENG IMPRINTED BY ESTRADIOL | 110 | 68 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR UP | 294 | 199 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER UP | 298 | 174 | All SZGR 2.0 genes in this pathway |
| FERRANDO HOX11 NEIGHBORS | 23 | 13 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS CTNNB1 DN | 170 | 105 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS POLYSOMY7 UP | 79 | 50 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 1HR DN | 222 | 147 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 3 UP | 170 | 97 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 6 UP | 140 | 81 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 7 UP | 118 | 68 | All SZGR 2.0 genes in this pathway |
| KIM TIAL1 TARGETS | 32 | 22 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
| SERVITJA LIVER HNF1A TARGETS DN | 157 | 105 | All SZGR 2.0 genes in this pathway |
| SMIRNOV RESPONSE TO IR 6HR UP | 166 | 97 | All SZGR 2.0 genes in this pathway |