| REACTOME DIABETES PATHWAYS
| 133 | 91 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 UP
| 408 | 247 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL DN
| 455 | 304 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN
| 460 | 312 | All SZGR 2.0 genes in this pathway |
| DOANE BREAST CANCER ESR1 DN
| 48 | 28 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER ADVANCED VS EARLY UP
| 175 | 120 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS UP
| 457 | 269 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP
| 501 | 327 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN
| 459 | 276 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR DN
| 129 | 84 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 12HR DN
| 209 | 122 | All SZGR 2.0 genes in this pathway |
| WATANABE ULCERATIVE COLITIS WITH CANCER DN
| 14 | 9 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED UP
| 633 | 376 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP
| 722 | 443 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS UP
| 149 | 84 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN
| 485 | 334 | All SZGR 2.0 genes in this pathway |
| VETTER TARGETS OF PRKCA AND ETS1 DN
| 16 | 10 | All SZGR 2.0 genes in this pathway |
| KAN RESPONSE TO ARSENIC TRIOXIDE
| 123 | 80 | All SZGR 2.0 genes in this pathway |
| WONG ENDMETRIUM CANCER UP
| 25 | 17 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER
| 747 | 453 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN
| 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN
| 483 | 336 | All SZGR 2.0 genes in this pathway |
| LIEN BREAST CARCINOMA METAPLASTIC VS DUCTAL UP
| 83 | 51 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK
| 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN
| 637 | 377 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS
| 539 | 324 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER E
| 89 | 44 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES 1
| 379 | 235 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS
| 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS
| 734 | 436 | All SZGR 2.0 genes in this pathway |
| MORI EMU MYC LYMPHOMA BY ONSET TIME UP
| 110 | 69 | All SZGR 2.0 genes in this pathway |
| PETROVA ENDOTHELIUM LYMPHATIC VS BLOOD DN
| 162 | 102 | All SZGR 2.0 genes in this pathway |
| PETROVA PROX1 TARGETS DN
| 64 | 38 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA DN
| 41 | 27 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 10HR DN
| 56 | 37 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS DN
| 371 | 218 | All SZGR 2.0 genes in this pathway |
| LEE AGING CEREBELLUM DN
| 86 | 66 | All SZGR 2.0 genes in this pathway |
| EHRLICH ICF SYNDROM UP
| 13 | 8 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR DN
| 101 | 70 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D6
| 37 | 25 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN
| 312 | 203 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK DN
| 318 | 220 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC UP
| 202 | 115 | All SZGR 2.0 genes in this pathway |
| DAZARD UV RESPONSE CLUSTER G6
| 153 | 112 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P6
| 91 | 44 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION DN
| 281 | 179 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE DN
| 315 | 197 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN
| 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP
| 648 | 398 | All SZGR 2.0 genes in this pathway |
| IZADPANAH STEM CELL ADIPOSE VS BONE DN
| 108 | 68 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR UP
| 294 | 199 | All SZGR 2.0 genes in this pathway |
| BREDEMEYER RAG SIGNALING VIA ATM NOT VIA NFKB UP
| 49 | 32 | All SZGR 2.0 genes in this pathway |
| LEE RECENT THYMIC EMIGRANT
| 227 | 128 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6
| 456 | 285 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K27ME3
| 269 | 159 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE UP
| 181 | 106 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G23 UP
| 52 | 35 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS PROLIFERATION UP
| 178 | 108 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED
| 351 | 238 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS DN
| 414 | 237 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN
| 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION DN
| 321 | 200 | All SZGR 2.0 genes in this pathway |
| SERVITJA LIVER HNF1A TARGETS UP
| 135 | 96 | All SZGR 2.0 genes in this pathway |
| WANG MLL TARGETS
| 289 | 188 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY
| 1839 | 928 | All SZGR 2.0 genes in this pathway |
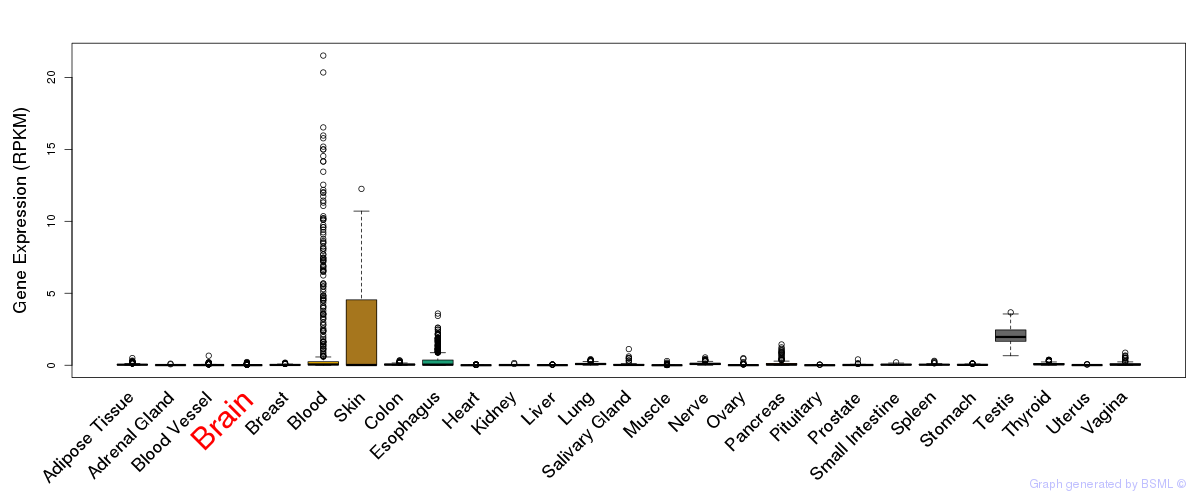
 Differentially methylated gene
Differentially methylated gene eQTL annotation
eQTL annotation