Gene Page: VCAN
Summary ?
| GeneID | 1462 |
| Symbol | VCAN |
| Synonyms | CSPG2|ERVR|GHAP|PG-M|WGN|WGN1 |
| Description | versican |
| Reference | MIM:118661|HGNC:HGNC:2464|Ensembl:ENSG00000038427|HPRD:00340|Vega:OTTHUMG00000131321 |
| Gene type | protein-coding |
| Map location | 5q14.3 |
| Pascal p-value | 0.037 |
| Sherlock p-value | 8.207E-4 |
| Fetal beta | 3.395 |
| DMG | 1 (# studies) |
| eGene | Cerebellum Myers' cis & trans |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| Expression | Meta-analysis of gene expression | P value: 3.122 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| VCAN | chr5 | 82837297 | A | G | NM_001126336 NM_001164097 NM_001164098 NM_004385 | . . . . | intronic silent intronic silent | Schizophrenia | DNM:Fromer_2014 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg00328900 | 5 | 82769097 | VCAN | 2.6E-8 | -0.022 | 8.31E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs11035999 | chr11 | 40679588 | VCAN | 1462 | 0.08 | trans |
Section II. Transcriptome annotation
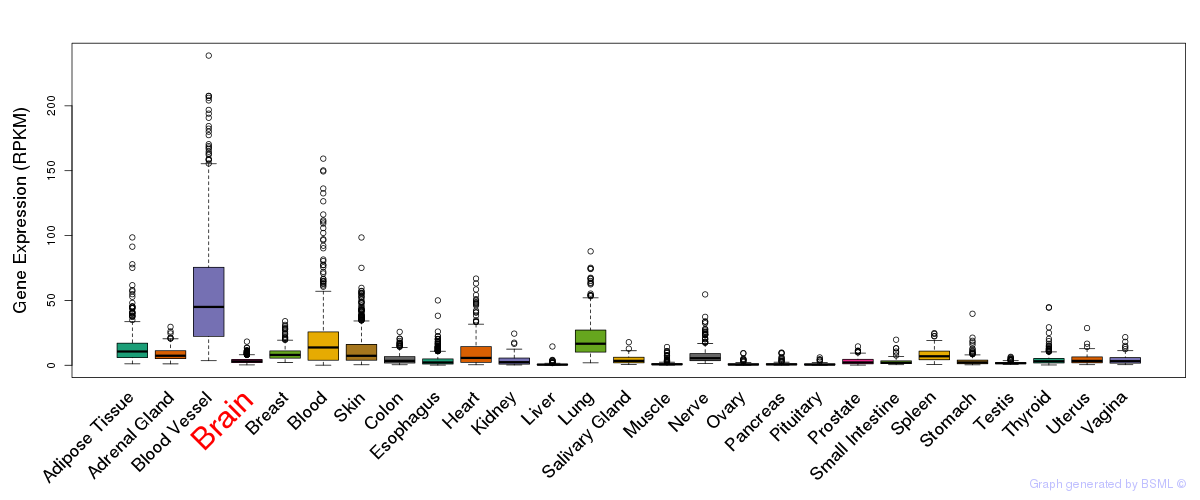
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005529 | sugar binding | IEA | - | |
| GO:0005540 | hyaluronic acid binding | TAS | 2583089 | |
| GO:0005488 | binding | IEA | - | |
| GO:0005509 | calcium ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007155 | cell adhesion | TAS | 2583089 | |
| GO:0007507 | heart development | IEA | - | |
| GO:0008037 | cell recognition | TAS | 2583089 | |
| GO:0007275 | multicellular organismal development | TAS | 2583089 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005578 | proteinaceous extracellular matrix | TAS | 2583089 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACAN | AGC1 | AGCAN | CSPG1 | CSPGCP | MSK16 | SEDK | aggrecan | Affinity Capture-Western Reconstituted Complex | BioGRID | 12888576 |
| CCL2 | GDCF-2 | HC11 | HSMCR30 | MCAF | MCP-1 | MCP1 | MGC9434 | SCYA2 | SMC-CF | chemokine (C-C motif) ligand 2 | - | HPRD,BioGRID | 11083865 |
| CCL20 | CKb4 | LARC | MIP-3a | MIP3A | SCYA20 | ST38 | chemokine (C-C motif) ligand 20 | - | HPRD,BioGRID | 11083865 |
| CCL21 | 6Ckine | CKb9 | ECL | MGC34555 | SCYA21 | SLC | TCA4 | chemokine (C-C motif) ligand 21 | - | HPRD,BioGRID | 11083865 |
| CCL5 | D17S136E | MGC17164 | RANTES | SCYA5 | SISd | TCP228 | chemokine (C-C motif) ligand 5 | - | HPRD,BioGRID | 11083865 |
| CCL8 | HC14 | MCP-2 | MCP2 | SCYA10 | SCYA8 | chemokine (C-C motif) ligand 8 | - | HPRD,BioGRID | 11083865 |
| CD44 | CDW44 | CSPG8 | ECMR-III | HCELL | IN | LHR | MC56 | MDU2 | MDU3 | MGC10468 | MIC4 | MUTCH-I | Pgp1 | CD44 molecule (Indian blood group) | - | HPRD,BioGRID | 10950950 |
| CXCL10 | C7 | IFI10 | INP10 | IP-10 | SCYB10 | crg-2 | gIP-10 | mob-1 | chemokine (C-X-C motif) ligand 10 | - | HPRD,BioGRID | 11083865 |
| FBLN1 | FBLN | fibulin 1 | - | HPRD,BioGRID | 10400671 |
| FBLN2 | - | fibulin 2 | - | HPRD,BioGRID | 11038354 |
| FBN1 | FBN | MASS | MFS1 | OCTD | SGS | WMS | fibrillin 1 | - | HPRD,BioGRID | 11726670 |
| HAPLN1 | CRTL1 | hyaluronan and proteoglycan link protein 1 | - | HPRD,BioGRID | 12888576 |
| ITGA4 | CD49D | IA4 | MGC90518 | integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) | - | HPRD,BioGRID | 11083865 |
| ITGB1 | CD29 | FNRB | GPIIA | MDF2 | MSK12 | VLA-BETA | VLAB | integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) | - | HPRD,BioGRID | 11805102 |
| PLA2G2A | MOM1 | PLA2 | PLA2B | PLA2L | PLA2S | PLAS1 | sPLA2 | phospholipase A2, group IIA (platelets, synovial fluid) | - | HPRD,BioGRID | 8824283 |9848887 |
| SELL | CD62L | LAM-1 | LAM1 | LECAM1 | LNHR | LSEL | LYAM1 | Leu-8 | Lyam-1 | PLNHR | TQ1 | hLHRc | selectin L | Reconstituted Complex | BioGRID | 10950950 |
| SELP | CD62 | CD62P | FLJ45155 | GMP140 | GRMP | LECAM3 | PADGEM | PSEL | selectin P (granule membrane protein 140kDa, antigen CD62) | - | HPRD,BioGRID | 10950950 |
| TNR | MGC149328 | TN-R | tenascin R (restrictin, janusin) | - | HPRD | 9294172 |
| XCL1 | ATAC | LPTN | LTN | SCM-1 | SCM-1a | SCM1 | SCYC1 | chemokine (C motif) ligand 1 | - | HPRD,BioGRID | 11083865 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CELL ADHESION MOLECULES CAMS | 134 | 93 | All SZGR 2.0 genes in this pathway |
| PID P53 DOWNSTREAM PATHWAY | 137 | 94 | All SZGR 2.0 genes in this pathway |
| PID BETA CATENIN NUC PATHWAY | 80 | 60 | All SZGR 2.0 genes in this pathway |
| PID TOLL ENDOGENOUS PATHWAY | 25 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME CS DS DEGRADATION | 12 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | 21 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | 49 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME HEPARAN SULFATE HEPARIN HS GAG METABOLISM | 52 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME GLYCOSAMINOGLYCAN METABOLISM | 111 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | 25 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF CARBOHYDRATES | 247 | 154 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL UP | 285 | 181 | All SZGR 2.0 genes in this pathway |
| LU TUMOR VASCULATURE UP | 29 | 17 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE UP | 351 | 230 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA UP | 294 | 178 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST DUCTAL CARCINOMA VS DUCTAL NORMAL UP | 44 | 25 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST DUCTAL CARCINOMA VS LOBULAR NORMAL UP | 73 | 50 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST LOBULAR CARCINOMA VS DUCTAL NORMAL UP | 69 | 38 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST LOBULAR CARCINOMA VS LOBULAR NORMAL DN | 74 | 42 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| LIU TARGETS OF VMYB VS CMYB DN | 43 | 30 | All SZGR 2.0 genes in this pathway |
| ROY WOUND BLOOD VESSEL UP | 50 | 30 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL DN | 244 | 153 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER ADVANCED VS EARLY UP | 175 | 120 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC UP | 185 | 126 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION ERYTHROCYTE UP | 157 | 104 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION SUSTAINDED IN ERYTHROCYTE UP | 44 | 30 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL DN | 226 | 132 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION CDC25 UP | 120 | 73 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 CHRONIC LOF UP | 115 | 78 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF DN | 228 | 137 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN DN | 172 | 112 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS G DN | 35 | 18 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK UP | 271 | 175 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 6 7WK DN | 79 | 54 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL TGFB1 TARGETS UP | 169 | 127 | All SZGR 2.0 genes in this pathway |
| LUCAS HNF4A TARGETS DN | 8 | 8 | All SZGR 2.0 genes in this pathway |
| KAN RESPONSE TO ARSENIC TRIOXIDE | 123 | 80 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER HIGH RECURRENCE | 49 | 31 | All SZGR 2.0 genes in this pathway |
| SCHAVOLT TARGETS OF TP53 AND TP63 | 16 | 12 | All SZGR 2.0 genes in this pathway |
| HERNANDEZ MITOTIC ARREST BY DOCETAXEL 2 DN | 19 | 13 | All SZGR 2.0 genes in this pathway |
| DACOSTA ERCC3 ALLELE XPCS VS TTD DN | 36 | 27 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS NEUROEPITHELIUM UP | 176 | 111 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 2 UP | 30 | 24 | All SZGR 2.0 genes in this pathway |
| INGRAM SHH TARGETS DN | 64 | 41 | All SZGR 2.0 genes in this pathway |
| MANTOVANI VIRAL GPCR SIGNALING DN | 49 | 37 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| LIAO HAVE SOX4 BINDING SITES | 40 | 26 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS | 539 | 324 | All SZGR 2.0 genes in this pathway |
| WU CELL MIGRATION | 184 | 114 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| SUNG METASTASIS STROMA UP | 110 | 70 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 UP | 256 | 159 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 SIGNALING VIA CTNNB1 | 83 | 58 | All SZGR 2.0 genes in this pathway |
| KANNAN TP53 TARGETS UP | 58 | 40 | All SZGR 2.0 genes in this pathway |
| WILLERT WNT SIGNALING | 24 | 13 | All SZGR 2.0 genes in this pathway |
| POMEROY MEDULLOBLASTOMA DESMOPLASIC VS CLASSIC DN | 59 | 35 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| HESS TARGETS OF HOXA9 AND MEIS1 DN | 77 | 48 | All SZGR 2.0 genes in this pathway |
| GERY CEBP TARGETS | 126 | 90 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED UP | 183 | 111 | All SZGR 2.0 genes in this pathway |
| PETROVA ENDOTHELIUM LYMPHATIC VS BLOOD DN | 162 | 102 | All SZGR 2.0 genes in this pathway |
| CHIARETTI ACUTE LYMPHOBLASTIC LEUKEMIA ZAP70 | 67 | 33 | All SZGR 2.0 genes in this pathway |
| PETROVA PROX1 TARGETS DN | 64 | 38 | All SZGR 2.0 genes in this pathway |
| HADDAD T LYMPHOCYTE AND NK PROGENITOR DN | 63 | 41 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP | 390 | 242 | All SZGR 2.0 genes in this pathway |
| WESTON VEGFA TARGETS 6HR | 59 | 38 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE UP | 249 | 170 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS UP | 37 | 27 | All SZGR 2.0 genes in this pathway |
| PAL PRMT5 TARGETS UP | 203 | 135 | All SZGR 2.0 genes in this pathway |
| JI RESPONSE TO FSH DN | 58 | 43 | All SZGR 2.0 genes in this pathway |
| MAHAJAN RESPONSE TO IL1A DN | 76 | 57 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| LINDVALL IMMORTALIZED BY TERT DN | 80 | 56 | All SZGR 2.0 genes in this pathway |
| WESTON VEGFA TARGETS | 108 | 71 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA RESPONSE TO TGFB1 C1 | 19 | 15 | All SZGR 2.0 genes in this pathway |
| GERHOLD ADIPOGENESIS DN | 64 | 44 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN UP | 262 | 186 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 3 | 101 | 64 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA EARLY RESPONSE TO TGFB1 | 58 | 43 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 24 HR DN | 91 | 64 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR DN | 178 | 121 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP | 487 | 303 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL DN | 128 | 93 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION DN | 287 | 208 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| BACOLOD RESISTANCE TO ALKYLATING AGENTS DN | 60 | 45 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS UP | 280 | 183 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN DN | 271 | 175 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS PEAK AT 16HR | 40 | 24 | All SZGR 2.0 genes in this pathway |
| BILD CTNNB1 ONCOGENIC SIGNATURE | 82 | 52 | All SZGR 2.0 genes in this pathway |
| CLASPER LYMPHATIC VESSELS DURING METASTASIS DN | 36 | 23 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE UP | 393 | 244 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 4 | 307 | 185 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN | 409 | 268 | All SZGR 2.0 genes in this pathway |
| SARRIO EPITHELIAL MESENCHYMAL TRANSITION UP | 180 | 114 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR DN | 191 | 123 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K9ME3 UP | 141 | 75 | All SZGR 2.0 genes in this pathway |
| HOQUE METHYLATED IN CANCER | 56 | 45 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL UP | 260 | 174 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| YAGUE PRETUMOR DRUG RESISTANCE DN | 13 | 11 | All SZGR 2.0 genes in this pathway |
| BRUECKNER TARGETS OF MIRLET7A3 UP | 111 | 69 | All SZGR 2.0 genes in this pathway |
| LU TUMOR ENDOTHELIAL MARKERS UP | 22 | 14 | All SZGR 2.0 genes in this pathway |
| LU TUMOR ANGIOGENESIS UP | 25 | 22 | All SZGR 2.0 genes in this pathway |
| ALONSO METASTASIS EMT UP | 36 | 24 | All SZGR 2.0 genes in this pathway |
| ALONSO METASTASIS UP | 198 | 128 | All SZGR 2.0 genes in this pathway |
| JIANG TIP30 TARGETS UP | 46 | 28 | All SZGR 2.0 genes in this pathway |
| WU SILENCED BY METHYLATION IN BLADDER CANCER | 55 | 42 | All SZGR 2.0 genes in this pathway |
| HELLEBREKERS SILENCED DURING TUMOR ANGIOGENESIS | 80 | 56 | All SZGR 2.0 genes in this pathway |
| TOOKER GEMCITABINE RESISTANCE DN | 122 | 84 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 DN | 315 | 201 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF DN | 235 | 144 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 DN | 245 | 150 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| CROONQUIST STROMAL STIMULATION UP | 60 | 42 | All SZGR 2.0 genes in this pathway |
| CROONQUIST NRAS VS STROMAL STIMULATION DN | 99 | 65 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 5 | 33 | 24 | All SZGR 2.0 genes in this pathway |
| POOLA INVASIVE BREAST CANCER UP | 288 | 168 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS CTNNB1 DN | 170 | 105 | All SZGR 2.0 genes in this pathway |
| LOPEZ MESOTHELIOMA SURVIVAL DN | 12 | 6 | All SZGR 2.0 genes in this pathway |
| KORKOLA EMBRYONIC CARCINOMA VS SEMINOMA UP | 22 | 15 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S1 | 237 | 159 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS UP | 221 | 135 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 16 | 79 | 47 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS DN | 229 | 135 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS DN | 315 | 215 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE VIA P38 COMPLETE | 227 | 151 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38 | 337 | 236 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
| PECE MAMMARY STEM CELL DN | 146 | 88 | All SZGR 2.0 genes in this pathway |
| ALTEMEIER RESPONSE TO LPS WITH MECHANICAL VENTILATION | 128 | 81 | All SZGR 2.0 genes in this pathway |
| ANASTASSIOU CANCER MESENCHYMAL TRANSITION SIGNATURE | 64 | 40 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |
| NABA PROTEOGLYCANS | 35 | 23 | All SZGR 2.0 genes in this pathway |
| NABA CORE MATRISOME | 275 | 148 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-101 | 293 | 299 | 1A | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-124/506 | 56 | 62 | m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-136 | 704 | 710 | m8 | hsa-miR-136 | ACUCCAUUUGUUUUGAUGAUGGA |
| miR-144 | 293 | 299 | 1A | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-186 | 238 | 244 | 1A | hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU |
| miR-203.1 | 30 | 36 | 1A | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| miR-496 | 333 | 340 | 1A,m8 | hsa-miR-496 | AUUACAUGGCCAAUCUC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.