Gene Page: EGFR
Summary ?
| GeneID | 1956 |
| Symbol | EGFR |
| Synonyms | ERBB|ERBB1|HER1|NISBD2|PIG61|mENA |
| Description | epidermal growth factor receptor |
| Reference | MIM:131550|HGNC:HGNC:3236|Ensembl:ENSG00000146648|HPRD:00579|Vega:OTTHUMG00000023661 |
| Gene type | protein-coding |
| Map location | 7p12 |
| Pascal p-value | 0.251 |
| Fetal beta | -0.918 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 1 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 2.8135 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg20706768 | 7 | 55225183 | EGFR | 4.973E-4 | 0.384 | 0.047 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
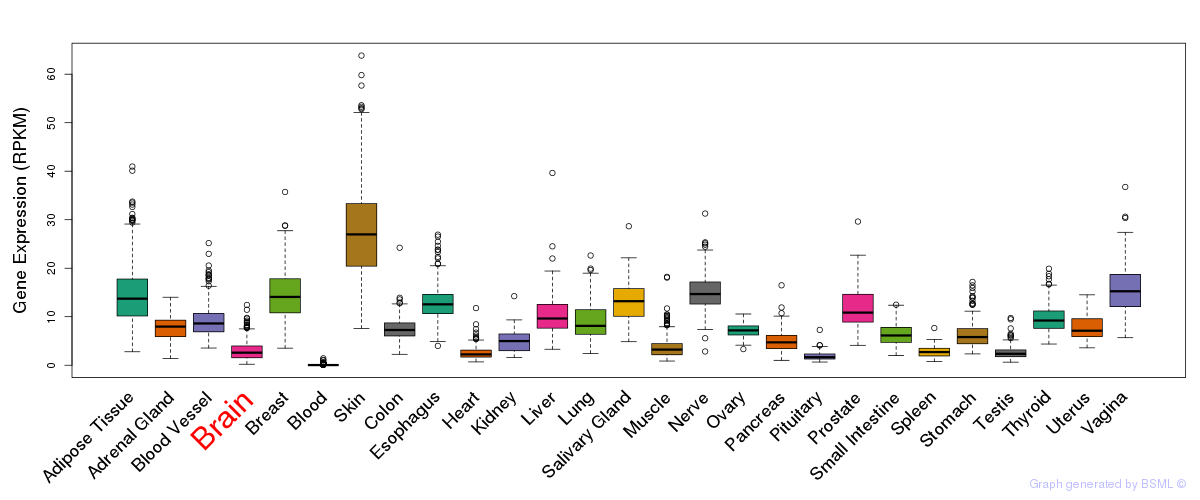
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NOTCH3 | 0.71 | 0.68 |
| SYDE1 | 0.70 | 0.69 |
| APLN | 0.69 | 0.62 |
| FGFR3 | 0.68 | 0.52 |
| NFKB2 | 0.68 | 0.67 |
| CASKIN2 | 0.68 | 0.65 |
| AC020763.2 | 0.68 | 0.68 |
| INPPL1 | 0.67 | 0.65 |
| SLC2A1 | 0.67 | 0.65 |
| PLXNB1 | 0.65 | 0.61 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TMEM70 | -0.47 | -0.48 |
| THOC7 | -0.47 | -0.52 |
| ZCCHC17 | -0.44 | -0.47 |
| GLMN | -0.44 | -0.48 |
| AP4S1 | -0.44 | -0.48 |
| C8orf38 | -0.44 | -0.46 |
| BEX5 | -0.44 | -0.47 |
| TCEAL7 | -0.42 | -0.49 |
| C2orf80 | -0.42 | -0.44 |
| MTIF3 | -0.42 | -0.48 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005006 | epidermal growth factor receptor activity | IDA | 12435727 | |
| GO:0005006 | epidermal growth factor receptor activity | NAS | 2790960 |6325948 | |
| GO:0004888 | transmembrane receptor activity | IDA | 7736574 | |
| GO:0003690 | double-stranded DNA binding | NAS | 6325948 | |
| GO:0030235 | nitric-oxide synthase regulator activity | IDA | 12828935 | |
| GO:0004710 | MAP/ERK kinase kinase activity | NAS | 15542601 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0051015 | actin filament binding | IDA | 14702346 | |
| GO:0042802 | identical protein binding | IPI | 16777603 | |
| GO:0046982 | protein heterodimerization activity | IDA | 10572067 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001503 | ossification | NAS | 12925580 | |
| GO:0007202 | activation of phospholipase C activity | TAS | 12435727 | |
| GO:0007173 | epidermal growth factor receptor signaling pathway | IDA | 12435727 | |
| GO:0006950 | response to stress | NAS | 12828935 | |
| GO:0007166 | cell surface receptor linked signal transduction | IDA | 7736574 | |
| GO:0016337 | cell-cell adhesion | IMP | 12435727 | |
| GO:0042327 | positive regulation of phosphorylation | IDA | 15082764 | |
| GO:0050999 | regulation of nitric-oxide synthase activity | IDA | 12828935 | |
| GO:0051205 | protein insertion into membrane | TAS | 12435727 | |
| GO:0030335 | positive regulation of cell migration | IMP | 12435727 | |
| GO:0043406 | positive regulation of MAP kinase activity | IDA | 10572067 | |
| GO:0043006 | calcium-dependent phospholipase A2 activation | TAS | 12435727 | |
| GO:0050679 | positive regulation of epithelial cell proliferation | IDA | 10572067 | |
| GO:0050730 | regulation of peptidyl-tyrosine phosphorylation | IMP | 12435727 | |
| GO:0045786 | negative regulation of cell cycle | IEA | - | |
| GO:0046777 | protein amino acid autophosphorylation | IEA | - | |
| GO:0045429 | positive regulation of nitric oxide biosynthetic process | IDA | 12828935 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005615 | extracellular space | NAS | 9103388 | |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005634 | nucleus | IDA | 12828935 | |
| GO:0005737 | cytoplasm | IDA | 12435727 | |
| GO:0005768 | endosome | IDA | 14702346 | |
| GO:0005783 | endoplasmic reticulum | IDA | 18029348 | |
| GO:0030122 | AP-2 adaptor complex | TAS | 14702346 | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005886 | plasma membrane | EXP | 8493579 |8639530 |8845374 |9356464 |9544989 |10567358 |10648629 |10734310 |10913131 |11823423 |11894095 |122181 | |
| GO:0005886 | plasma membrane | IDA | 15465819 | |
| GO:0016323 | basolateral plasma membrane | IEA | - | |
| GO:0030139 | endocytic vesicle | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AMH | MIF | MIS | anti-Mullerian hormone | - | HPRD | 8596488 |
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | Affinity Capture-Western Co-localization | BioGRID | 15288768 |15305378 |
| AREG | AR | CRDGF | MGC13647 | SDGF | amphiregulin | - | HPRD,BioGRID | 10085134 |
| ARF4 | ARF2 | ADP-ribosylation factor 4 | - | HPRD,BioGRID | 12446727 |
| ATP5C1 | ATP5C | ATP5CL1 | ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 | Two-hybrid | BioGRID | 16169070 |
| BTC | - | betacellulin | - | HPRD | 9528863 |
| CALM1 | CALML2 | CAMI | DD132 | PHKD | calmodulin 1 (phosphorylase kinase, delta) | EGFR interacts with calmodulin | BIND | 14960328 |
| CAMLG | CAML | MGC163197 | calcium modulating ligand | - | HPRD,BioGRID | 12919676 |
| CASP1 | ICE | IL1BC | P45 | caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase) | - | HPRD,BioGRID | 11226410 |
| CAV1 | CAV | MSTP085 | VIP21 | caveolin 1, caveolae protein, 22kDa | - | HPRD,BioGRID | 9374534 |
| CAV3 | LGMD1C | LQT9 | MGC126100 | MGC126101 | MGC126129 | VIP-21 | VIP21 | caveolin 3 | - | HPRD,BioGRID | 9374534 |
| CBL | C-CBL | CBL2 | RNF55 | Cas-Br-M (murine) ecotropic retroviral transforming sequence | Affinity Capture-Western | BioGRID | 10086340 |12061819 |
| CBL | C-CBL | CBL2 | RNF55 | Cas-Br-M (murine) ecotropic retroviral transforming sequence | - | HPRD | 8662998 |
| CBL | C-CBL | CBL2 | RNF55 | Cas-Br-M (murine) ecotropic retroviral transforming sequence | An unspecified isoform of EGFR interacts with CBL. | BIND | 15735736 |
| CBL | C-CBL | CBL2 | RNF55 | Cas-Br-M (murine) ecotropic retroviral transforming sequence | c-Cbl ubiquitinates an unspecified isoform of EGFR. This interaction was modeled on a demonstrated interaction between c-Cbl and EGFR, both from an unspecified species. | BIND | 12234920 |
| CBLB | DKFZp686J10223 | DKFZp779A0729 | DKFZp779F1443 | FLJ36865 | FLJ41152 | Nbla00127 | RNF56 | Cas-Br-M (murine) ecotropic retroviral transforming sequence b | - | HPRD,BioGRID | 10086340 |
| CBLC | CBL-3 | CBL-SL | RNF57 | Cas-Br-M (murine) ecotropic retroviral transforming sequence c | - | HPRD,BioGRID | 10362357 |
| CCND1 | BCL1 | D11S287E | PRAD1 | U21B31 | cyclin D1 | An unspecified isoform of EGFR binds to Cyclin D1 promoter. | BIND | 15950906 |
| CD44 | CDW44 | CSPG8 | ECMR-III | HCELL | IN | LHR | MC56 | MDU2 | MDU3 | MGC10468 | MIC4 | MUTCH-I | Pgp1 | CD44 molecule (Indian blood group) | CD44 interacts with EGFR. | BIND | 12093135 |
| CD44 | CDW44 | CSPG8 | ECMR-III | HCELL | IN | LHR | MC56 | MDU2 | MDU3 | MGC10468 | MIC4 | MUTCH-I | Pgp1 | CD44 molecule (Indian blood group) | - | HPRD,BioGRID | 12093135 |
| CD82 | 4F9 | C33 | GR15 | IA4 | KAI1 | R2 | SAR2 | ST6 | TSPAN27 | CD82 molecule | - | HPRD,BioGRID | 10985391 |
| CDC25A | CDC25A2 | cell division cycle 25 homolog A (S. pombe) | - | HPRD,BioGRID | 11912208 |
| CDH1 | Arc-1 | CD324 | CDHE | ECAD | LCAM | UVO | cadherin 1, type 1, E-cadherin (epithelial) | EGFR interacts with E-cadherin | BIND | 14681060 |
| CDH1 | Arc-1 | CD324 | CDHE | ECAD | LCAM | UVO | cadherin 1, type 1, E-cadherin (epithelial) | - | HPRD,BioGRID | 10969083 |
| CEBPB | C/EBP-beta | CRP2 | IL6DBP | LAP | MGC32080 | NF-IL6 | TCF5 | CCAAT/enhancer binding protein (C/EBP), beta | - | HPRD,BioGRID | 12095417 |
| CRK | CRKII | v-crk sarcoma virus CT10 oncogene homolog (avian) | - | HPRD,BioGRID | 9642287 |
| CSK | MGC117393 | c-src tyrosine kinase | Affinity Capture-MS | BioGRID | 16729043 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | - | HPRD,BioGRID | 9233779 |
| CTNND1 | CAS | CTNND | KIAA0384 | P120CAS | P120CTN | p120 | catenin (cadherin-associated protein), delta 1 | Reconstituted Complex | BioGRID | 9535896 |
| DCN | CSCD | DSPG2 | PG40 | PGII | PGS2 | SLRR1B | decorin | - | HPRD,BioGRID | 9988678 |12105206 |
| DEGS1 | DEGS | DES1 | Des-1 | FADS7 | MGC5079 | MIG15 | MLD | degenerative spermatocyte homolog 1, lipid desaturase (Drosophila) | - | HPRD | 9188692 |
| DOK2 | p56DOK | p56dok-2 | docking protein 2, 56kDa | - | HPRD,BioGRID | 10508618 |
| DUSP3 | VHR | dual specificity phosphatase 3 | Biochemical Activity | BioGRID | 1281549 |
| EGF | HOMG4 | URG | epidermal growth factor (beta-urogastrone) | - | HPRD | 10788520 |12093292 |12620237|12093292 |
| EGF | HOMG4 | URG | epidermal growth factor (beta-urogastrone) | EGF interacts with EGFR. | BIND | 15837620 |
| EGF | HOMG4 | URG | epidermal growth factor (beta-urogastrone) | - | HPRD,BioGRID | 12093292 |
| EGF | HOMG4 | URG | epidermal growth factor (beta-urogastrone) | EGFR interacts with EGF. This interaction was modelled on a demonstrated interaction between EGFR from human and EGF from mouse. | BIND | 8413296 |
| EGF | HOMG4 | URG | epidermal growth factor (beta-urogastrone) | An unspecified isoform of EGFR interacts with EGF. | BIND | 15950906 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | EGFR interacts with EGFR. | BIND | 15284455 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | Affinity Capture-Western | BioGRID | 14576349 |
| EPPK1 | EPIPL | EPIPL1 | epiplakin 1 | - | HPRD | 12577067 |
| EPS15 | AF-1P | AF1P | MLLT5 | epidermal growth factor receptor pathway substrate 15 | - | HPRD,BioGRID | 9049247 |
| EPS8 | - | epidermal growth factor receptor pathway substrate 8 | - | HPRD | 7532293 |12127568 |
| ERBB2 | CD340 | HER-2 | HER-2/neu | HER2 | NEU | NGL | TKR1 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) | - | HPRD | 11500850 |
| ERBB2 | CD340 | HER-2 | HER-2/neu | HER2 | NEU | NGL | TKR1 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) | EGFR interacts with ErbB2. | BIND | 15652750 |
| ERBB3 | ErbB-3 | HER3 | LCCS2 | MDA-BF-1 | MGC88033 | c-erbB-3 | c-erbB3 | erbB3-S | p180-ErbB3 | p45-sErbB3 | p85-sErbB3 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian) | - | HPRD,BioGRID | 10527633 |
| EREG | ER | epiregulin | - | HPRD,BioGRID | 9419975 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | - | HPRD,BioGRID | 11887937 |
| FER | TYK3 | fer (fps/fes related) tyrosine kinase | - | HPRD,BioGRID | 7623846 |
| FOS | AP-1 | C-FOS | v-fos FBJ murine osteosarcoma viral oncogene homolog | An unspecified isoform of EGFR binds to c-fos promoter. | BIND | 15950906 |
| GAB1 | - | GRB2-associated binding protein 1 | - | HPRD,BioGRID | 11432805 |
| GNAI2 | GIP | GNAI2B | H_LUCA15.1 | H_LUCA16.1 | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 | - | HPRD | 11286993 |
| GRB10 | GRB-IR | Grb-10 | IRBP | KIAA0207 | MEG1 | RSS | growth factor receptor-bound protein 10 | Grb10 interacts with EGFR. | BIND | 9506989 |
| GRB10 | GRB-IR | Grb-10 | IRBP | KIAA0207 | MEG1 | RSS | growth factor receptor-bound protein 10 | - | HPRD,BioGRID | 9006901 |9506989 |
| GRB14 | - | growth factor receptor-bound protein 14 | - | HPRD,BioGRID | 8647858 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | GRB2 interacts with an unspecified isoform of EGFR. This interaction was modeled on a demonstrated interaction between human GRB2, and rat EGFR. | BIND | 15782189 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | - | HPRD | 1322798 |7527043 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | EGFR interacts with GRB2. | BIND | 8305738 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | Affinity Capture-MS Affinity Capture-Western Reconstituted Complex | BioGRID | 1322798 |7510700 |7527043 |8647858 |9050991 |10026169 |10085134 |11960376 |12577067 |16729043 |
| GRB7 | - | growth factor receptor-bound protein 7 | - | HPRD | 9710451 |
| HBEGF | DTR | DTS | DTSF | HEGFL | heparin-binding EGF-like growth factor | - | HPRD,BioGRID | 12725245 |
| HSP90AA1 | FLJ31884 | HSP86 | HSP89A | HSP90A | HSP90N | HSPC1 | HSPCA | HSPCAL1 | HSPCAL4 | HSPN | Hsp89 | Hsp90 | LAP2 | heat shock protein 90kDa alpha (cytosolic), class A member 1 | EGFR interacts with Hsp90. This interaction was modeled on a demonstrated interaction between EGFR from an unspecified species and human Hsp90. | BIND | 12471035 |
| HTT | HD | IT15 | huntingtin | - | HPRD,BioGRID | 9079622 |
| IGF1R | CD221 | IGFIR | JTK13 | MGC142170 | MGC142172 | MGC18216 | insulin-like growth factor 1 receptor | Affinity Capture-Western | BioGRID | 17047074 |
| INPPL1 | SHIP2 | inositol polyphosphate phosphatase-like 1 | - | HPRD,BioGRID | 11349134 |
| ITGA5 | CD49e | FNRA | VLA5A | integrin, alpha 5 (fibronectin receptor, alpha polypeptide) | - | HPRD | 10888683 |
| JAK2 | JTK10 | Janus kinase 2 (a protein tyrosine kinase) | Affinity Capture-Western | BioGRID | 10358079 |
| JUP | ARVD12 | CTNNG | DP3 | DPIII | PDGB | PKGB | junction plakoglobin | Reconstituted Complex | BioGRID | 9535896 |
| KRT17 | K17 | PC | PC2 | PCHC1 | keratin 17 | - | HPRD | 12577067 |
| KRT18 | CYK18 | K18 | keratin 18 | - | HPRD | 12577067 |
| KRT7 | CK7 | K2C7 | K7 | MGC129731 | MGC3625 | SCL | keratin 7 | - | HPRD | 12577067 |
| KRT8 | CARD2 | CK8 | CYK8 | K2C8 | K8 | KO | keratin 8 | - | HPRD | 12577067 |
| LRSAM1 | FLJ31641 | RIFLE | TAL | leucine rich repeat and sterile alpha motif containing 1 | Affinity Capture-Western | BioGRID | 15256501 |
| MAP3K14 | FTDCR1B | HS | HSNIK | NIK | mitogen-activated protein kinase kinase kinase 14 | - | HPRD,BioGRID | 11116146 |
| MAP4K1 | HPK1 | mitogen-activated protein kinase kinase kinase kinase 1 | - | HPRD,BioGRID | 9346925 |
| MAPK14 | CSBP1 | CSBP2 | CSPB1 | EXIP | Mxi2 | PRKM14 | PRKM15 | RK | SAPK2A | p38 | p38ALPHA | mitogen-activated protein kinase 14 | Biochemical Activity | BioGRID | 7535770 |
| MUC1 | CD227 | EMA | H23AG | MAM6 | PEM | PEMT | PUM | mucin 1, cell surface associated | - | HPRD,BioGRID | 11278868 |11483589 |
| MUC1 | CD227 | EMA | H23AG | MAM6 | PEM | PEMT | PUM | mucin 1, cell surface associated | MUC1 interacts with and is tyrosine-phosphorylated by EGFR. | BIND | 11483589 |
| NCK1 | MGC12668 | NCK | NCKalpha | NCK adaptor protein 1 | - | HPRD,BioGRID | 9362449 |
| NCK2 | GRB4 | NCKbeta | NCK adaptor protein 2 | Affinity Capture-Western Reconstituted Complex | BioGRID | 9737977 |10026169 |
| NCK2 | GRB4 | NCKbeta | NCK adaptor protein 2 | - | HPRD | 1333047 |9737977 |
| NRG1 | ARIA | GGF | GGF2 | HGL | HRG | HRG1 | HRGA | NDF | SMDF | neuregulin 1 | Affinity Capture-Western | BioGRID | 7730382 |
| NRG1 | ARIA | GGF | GGF2 | HGL | HRG | HRG1 | HRGA | NDF | SMDF | neuregulin 1 | - | HPRD | 8702572 |
| PAK1 | MGC130000 | MGC130001 | PAKalpha | p21 protein (Cdc42/Rac)-activated kinase 1 | Tyrosine-phosphorylated EGFR interacts with PAK1. | BIND | 12522133 |
| PDGFRB | CD140B | JTK12 | PDGF-R-beta | PDGFR | PDGFR1 | platelet-derived growth factor receptor, beta polypeptide | - | HPRD,BioGRID | 9506992 |
| PIK3C2B | C2-PI3K | DKFZp686G16234 | phosphoinositide-3-kinase, class 2, beta polypeptide | - | HPRD,BioGRID | 11533253 |
| PIK3R1 | GRB1 | p85 | p85-ALPHA | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | - | HPRD | 8662998 |
| PIK3R1 | GRB1 | p85 | p85-ALPHA | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | EGFR interacts with p85. This interaction was modeled on a demonstrated interaction between human EGFR and bovine p85. | BIND | 1372092 |
| PIK3R2 | P85B | p85 | p85-BETA | phosphoinositide-3-kinase, regulatory subunit 2 (beta) | Tyrosine phosphorlyated EGFR interacts with p85-beta. | BIND | 11172806 |
| PITPNA | MGC99649 | PITPN | VIB1A | phosphatidylinositol transfer protein, alpha | - | HPRD,BioGRID | 7761838 |
| PLCG1 | PLC-II | PLC1 | PLC148 | PLCgamma1 | phospholipase C, gamma 1 | - | HPRD,BioGRID | 9207933 |
| PLCG1 | PLC-II | PLC1 | PLC148 | PLCgamma1 | phospholipase C, gamma 1 | EGFR interacts with PLC-gamma. This interaction was modelled on a demonstrated interaction between human EGFR and bovine PLC-gamma. | BIND | 2173144 |2176151 |
| PLD2 | - | phospholipase D2 | - | HPRD,BioGRID | 9837959 |
| PLD2 | - | phospholipase D2 | EGFR interacts with PLD1. This interaction was modelled on a demonstrated interaction between human EGFR and mouse PLD2. | BIND | 9837959 |
| PLEC1 | EBS1 | EBSO | HD1 | PCN | PLEC1b | PLTN | plectin 1, intermediate filament binding protein 500kDa | - | HPRD | 12577067 |
| PLSCR1 | MMTRA1B | phospholipid scramblase 1 | - | HPRD,BioGRID | 12009895 |
| PLSCR1 | MMTRA1B | phospholipid scramblase 1 | - | HPRD | 12009895 |12871937 |
| PRKACA | MGC102831 | MGC48865 | PKACA | protein kinase, cAMP-dependent, catalytic, alpha | - | HPRD,BioGRID | 9050991 |
| PRKAR1A | CAR | CNC | CNC1 | DKFZp779L0468 | MGC17251 | PKR1 | PPNAD1 | PRKAR1 | TSE1 | protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) | - | HPRD,BioGRID | 9050991 |
| PRKCA | AAG6 | MGC129900 | MGC129901 | PKC-alpha | PKCA | PRKACA | protein kinase C, alpha | - | HPRD,BioGRID | 12878187 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | - | HPRD,BioGRID | 10806474 |
| PTK2B | CADTK | CAKB | FADK2 | FAK2 | FRNK | PKB | PTK | PYK2 | RAFTK | PTK2B protein tyrosine kinase 2 beta | - | HPRD,BioGRID | 10777553 |
| PTK6 | BRK | FLJ42088 | PTK6 protein tyrosine kinase 6 | - | HPRD,BioGRID | 8940083 |
| PTPN1 | PTP1B | protein tyrosine phosphatase, non-receptor type 1 | - | HPRD,BioGRID | 7540771 |8621392 |10889023 |12573287 |
| PTPN11 | BPTP3 | CFC | MGC14433 | NS1 | PTP-1D | PTP2C | SH-PTP2 | SH-PTP3 | SHP2 | protein tyrosine phosphatase, non-receptor type 11 | - | HPRD,BioGRID | 7673163 |
| PTPN6 | HCP | HCPH | HPTP1C | PTP-1C | SH-PTP1 | SHP-1 | SHP-1L | SHP1 | protein tyrosine phosphatase, non-receptor type 6 | - | HPRD,BioGRID | 9733788 |
| PTPRJ | CD148 | DEP1 | HPTPeta | R-PTP-ETA | SCC1 | protein tyrosine phosphatase, receptor type, J | - | HPRD | 9115287 |
| RASA1 | CM-AVM | CMAVM | DKFZp434N071 | GAP | PKWS | RASA | RASGAP | p120GAP | p120RASGAP | RAS p21 protein activator (GTPase activating protein) 1 | Reconstituted Complex | BioGRID | 8647858 |
| RASA1 | CM-AVM | CMAVM | DKFZp434N071 | GAP | PKWS | RASA | RASGAP | p120GAP | p120RASGAP | RAS p21 protein activator (GTPase activating protein) 1 | GAP interacts with EGFR. | BIND | 1385407 |1850098 |2176151 |
| RASA1 | CM-AVM | CMAVM | DKFZp434N071 | GAP | PKWS | RASA | RASGAP | p120GAP | p120RASGAP | RAS p21 protein activator (GTPase activating protein) 1 | - | HPRD | 1633149 |
| RGS16 | A28-RGS14 | A28-RGS14P | RGS-R | regulator of G-protein signaling 16 | - | HPRD,BioGRID | 11602604 |
| RIPK1 | FLJ39204 | RIP | RIP1 | receptor (TNFRSF)-interacting serine-threonine kinase 1 | - | HPRD,BioGRID | 11116146 |
| ROS1 | MCF3 | ROS | c-ros-1 | c-ros oncogene 1 , receptor tyrosine kinase | Affinity Capture-Western | BioGRID | 11094073 |
| SGSM2 | KIAA0397 | RUTBC1 | small G protein signaling modulator 2 | Two-hybrid | BioGRID | 16169070 |
| SH2D3A | NSP1 | SH2 domain containing 3A | - | HPRD,BioGRID | 10187783 |
| SH3BGRL | MGC117402 | SH3BGR | SH3 domain binding glutamic acid-rich protein like | Affinity Capture-MS | BioGRID | 16729043 |
| SH3KBP1 | CIN85 | GIG10 | MIG18 | SH3-domain kinase binding protein 1 | Affinity Capture-Western | BioGRID | 11894095 |12177062 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | Shc interacts with activated EGFR. | BIND | 1623525 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | ShcA interacts with EGFR. | BIND | 8610109 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | ShcA interacts with the EGFR. | BIND | 8577769 |8610109 |8943228 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | - | HPRD,BioGRID | 9544989 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | EGF-R interacts with Shc. This interaction was modelled on a demonstrated interaction between human EGF-R and mouse Shc. | BIND | 8887653 |
| SHC3 | N-Shc | NSHC | RAI | SHCC | SHC (Src homology 2 domain containing) transforming protein 3 | - | HPRD,BioGRID | 9507002 |
| SNRPD2 | SMD2 | SNRPD1 | small nuclear ribonucleoprotein D2 polypeptide 16.5kDa | - | HPRD | 12577067 |
| SNX1 | HsT17379 | MGC8664 | SNX1A | Vps5 | sorting nexin 1 | - | HPRD,BioGRID | 8638121 |9819414 |
| SNX2 | MGC5204 | TRG-9 | sorting nexin 2 | - | HPRD,BioGRID | 9819414 |
| SNX4 | - | sorting nexin 4 | - | HPRD,BioGRID | 9819414 |
| SNX6 | MGC3157 | MSTP010 | TFAF2 | sorting nexin 6 | - | HPRD,BioGRID | 11279102 |
| SOCS1 | CIS1 | CISH1 | JAB | SOCS-1 | SSI-1 | SSI1 | TIP3 | suppressor of cytokine signaling 1 | - | HPRD,BioGRID | 12070153 |
| SOCS3 | ATOD4 | CIS3 | Cish3 | MGC71791 | SOCS-3 | SSI-3 | SSI3 | suppressor of cytokine signaling 3 | - | HPRD,BioGRID | 12070153 |
| SOS1 | GF1 | GGF1 | GINGF | HGF | NS4 | son of sevenless homolog 1 (Drosophila) | - | HPRD,BioGRID | 9447973 |
| SOS2 | FLJ25596 | son of sevenless homolog 2 (Drosophila) | - | HPRD,BioGRID | 10675333 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | - | HPRD,BioGRID | 10971656 |
| STAT1 | DKFZp686B04100 | ISGF-3 | STAT91 | signal transducer and activator of transcription 1, 91kDa | - | HPRD,BioGRID | 12070153 |
| STAT3 | APRF | FLJ20882 | HIES | MGC16063 | signal transducer and activator of transcription 3 (acute-phase response factor) | Affinity Capture-Western | BioGRID | 10358079 |15485908 |
| STAT3 | APRF | FLJ20882 | HIES | MGC16063 | signal transducer and activator of transcription 3 (acute-phase response factor) | Tyrosine phophorylated EGFR interacts with STAT3. | BIND | 12873986 |
| STAT5A | MGF | STAT5 | signal transducer and activator of transcription 5A | - | HPRD,BioGRID | 10358079 |
| STAT5B | STAT5 | signal transducer and activator of transcription 5B | - | HPRD,BioGRID | 10558875 |
| TGFA | TFGA | transforming growth factor, alpha | - | HPRD,BioGRID | 12297049 |
| TJP1 | DKFZp686M05161 | MGC133289 | ZO-1 | tight junction protein 1 (zona occludens 1) | - | HPRD,BioGRID | 12708492 |
| TNC | HXB | MGC167029 | TN | tenascin C | - | HPRD | 11470832 |
| TNK2 | ACK | ACK1 | FLJ44758 | FLJ45547 | p21cdc42Hs | tyrosine kinase, non-receptor, 2 | - | HPRD | 8497321 |8647288 |
| VAV1 | VAV | vav 1 guanine nucleotide exchange factor | - | HPRD,BioGRID | 10938113 |
| VAV2 | - | vav 2 guanine nucleotide exchange factor | Tyrosine phosphorylated EGFR interacts with and phosphorylates Vav2. | BIND | 12454019 |
| VAV2 | - | vav 2 guanine nucleotide exchange factor | - | HPRD,BioGRID | 10618391 |10938113 |
| VAV3 | FLJ40431 | vav 3 guanine nucleotide exchange factor | Affinity Capture-Western | BioGRID | 10938113 |
| VAV3 | FLJ40431 | vav 3 guanine nucleotide exchange factor | - | HPRD | 11094073 |
| WAS | IMD2 | THC | WASP | Wiskott-Aldrich syndrome (eczema-thrombocytopenia) | Affinity Capture-Western | BioGRID | 9307968 |
| XPO6 | EXP6 | FLJ22519 | KIAA0370 | RANBP20 | exportin 6 | - | HPRD | 14592989 |
| XRCC6 | CTC75 | CTCBF | G22P1 | KU70 | ML8 | TLAA | X-ray repair complementing defective repair in Chinese hamster cells 6 | - | HPRD | 9430697 |
| YWHAZ | KCIP-1 | MGC111427 | MGC126532 | MGC138156 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | EGFR interacts with 14-3-3-zeta. | BIND | 15225635 |
| ZNF259 | MGC110983 | ZPR1 | zinc finger protein 259 | - | HPRD | 8650580 |10938113|10938113 |
| ZNF259 | MGC110983 | ZPR1 | zinc finger protein 259 | - | HPRD | 10938113 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG ERBB SIGNALING PATHWAY | 87 | 71 | All SZGR 2.0 genes in this pathway |
| KEGG CALCIUM SIGNALING PATHWAY | 178 | 134 | All SZGR 2.0 genes in this pathway |
| KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION | 267 | 161 | All SZGR 2.0 genes in this pathway |
| KEGG ENDOCYTOSIS | 183 | 132 | All SZGR 2.0 genes in this pathway |
| KEGG DORSO VENTRAL AXIS FORMATION | 25 | 21 | All SZGR 2.0 genes in this pathway |
| KEGG FOCAL ADHESION | 201 | 138 | All SZGR 2.0 genes in this pathway |
| KEGG ADHERENS JUNCTION | 75 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG GAP JUNCTION | 90 | 68 | All SZGR 2.0 genes in this pathway |
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| KEGG GNRH SIGNALING PATHWAY | 101 | 77 | All SZGR 2.0 genes in this pathway |
| KEGG EPITHELIAL CELL SIGNALING IN HELICOBACTER PYLORI INFECTION | 68 | 44 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG PANCREATIC CANCER | 70 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG ENDOMETRIAL CANCER | 52 | 45 | All SZGR 2.0 genes in this pathway |
| KEGG GLIOMA | 65 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG PROSTATE CANCER | 89 | 75 | All SZGR 2.0 genes in this pathway |
| KEGG MELANOMA | 71 | 57 | All SZGR 2.0 genes in this pathway |
| KEGG BLADDER CANCER | 42 | 33 | All SZGR 2.0 genes in this pathway |
| KEGG NON SMALL CELL LUNG CANCER | 54 | 47 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AGR PATHWAY | 36 | 31 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AT1R PATHWAY | 36 | 28 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CBL PATHWAY | 13 | 10 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EGF PATHWAY | 31 | 25 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ERK PATHWAY | 28 | 22 | All SZGR 2.0 genes in this pathway |
| BIOCARTA KERATINOCYTE PATHWAY | 46 | 38 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EGFR SMRTE PATHWAY | 11 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MCALPAIN PATHWAY | 25 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CARDIACEGF PATHWAY | 18 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HER2 PATHWAY | 22 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA SPRY PATHWAY | 18 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TEL PATHWAY | 18 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TFF PATHWAY | 21 | 15 | All SZGR 2.0 genes in this pathway |
| ST G ALPHA I PATHWAY | 35 | 29 | All SZGR 2.0 genes in this pathway |
| ST ADRENERGIC | 36 | 29 | All SZGR 2.0 genes in this pathway |
| ST FAS SIGNALING PATHWAY | 65 | 54 | All SZGR 2.0 genes in this pathway |
| PID LYSOPHOSPHOLIPID PATHWAY | 66 | 53 | All SZGR 2.0 genes in this pathway |
| PID PTP1B PATHWAY | 52 | 40 | All SZGR 2.0 genes in this pathway |
| PID ARF6 PATHWAY | 35 | 27 | All SZGR 2.0 genes in this pathway |
| PID TCPTP PATHWAY | 43 | 33 | All SZGR 2.0 genes in this pathway |
| PID TXA2PATHWAY | 57 | 43 | All SZGR 2.0 genes in this pathway |
| PID SHP2 PATHWAY | 58 | 46 | All SZGR 2.0 genes in this pathway |
| PID TELOMERASE PATHWAY | 68 | 48 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 RECEPTOR PROXIMAL PATHWAY | 35 | 29 | All SZGR 2.0 genes in this pathway |
| PID AJDISS 2PATHWAY | 48 | 38 | All SZGR 2.0 genes in this pathway |
| PID P53 DOWNSTREAM PATHWAY | 137 | 94 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 DOWNSTREAM PATHWAY | 105 | 78 | All SZGR 2.0 genes in this pathway |
| PID UPA UPAR PATHWAY | 42 | 30 | All SZGR 2.0 genes in this pathway |
| PID ECADHERIN KERATINOCYTE PATHWAY | 21 | 19 | All SZGR 2.0 genes in this pathway |
| PID ERBB NETWORK PATHWAY | 15 | 13 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 INTERNALIZATION PATHWAY | 41 | 35 | All SZGR 2.0 genes in this pathway |
| PID ECADHERIN STABILIZATION PATHWAY | 42 | 34 | All SZGR 2.0 genes in this pathway |
| PID A6B1 A6B4 INTEGRIN PATHWAY | 46 | 35 | All SZGR 2.0 genes in this pathway |
| PID SYNDECAN 3 PATHWAY | 17 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB4 | 90 | 67 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB2 | 101 | 78 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | 18 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | 22 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY EGFR IN CANCER | 109 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K EVENTS IN ERBB2 SIGNALING | 44 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME GASTRIN CREB SIGNALLING PATHWAY VIA PKC AND MAPK | 205 | 136 | All SZGR 2.0 genes in this pathway |
| REACTOME EGFR DOWNREGULATION | 25 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME GAB1 SIGNALOSOME | 38 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME SHC1 EVENTS IN EGFR SIGNALING | 15 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME AXON GUIDANCE | 251 | 188 | All SZGR 2.0 genes in this pathway |
| REACTOME L1CAM INTERACTIONS | 86 | 62 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNAL TRANSDUCTION BY L1 | 34 | 25 | All SZGR 2.0 genes in this pathway |
| LIU SOX4 TARGETS UP | 137 | 94 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS DN | 182 | 111 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST DUCTAL CARCINOMA VS DUCTAL NORMAL DN | 198 | 110 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST LOBULAR CARCINOMA VS LOBULAR NORMAL UP | 94 | 59 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL DN | 455 | 304 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| DOANE BREAST CANCER ESR1 DN | 48 | 28 | All SZGR 2.0 genes in this pathway |
| NOJIMA SFRP2 TARGETS DN | 25 | 18 | All SZGR 2.0 genes in this pathway |
| JAEGER METASTASIS DN | 258 | 141 | All SZGR 2.0 genes in this pathway |
| PAPASPYRIDONOS UNSTABLE ATEROSCLEROTIC PLAQUE DN | 43 | 29 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS UP | 238 | 144 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA UP | 544 | 308 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 12HR DN | 209 | 122 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA UP | 171 | 112 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA BY DMOG UP | 130 | 85 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HIF1A TARGETS DN | 91 | 58 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HIF1A AND HIF2A TARGETS DN | 104 | 72 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| PROVENZANI METASTASIS DN | 136 | 94 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 CHRONIC LOF DN | 118 | 78 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS F UP | 185 | 119 | All SZGR 2.0 genes in this pathway |
| OZANNE AP1 TARGETS UP | 18 | 11 | All SZGR 2.0 genes in this pathway |
| APPIERTO RESPONSE TO FENRETINIDE DN | 51 | 35 | All SZGR 2.0 genes in this pathway |
| BREUHAHN GROWTH FACTOR SIGNALING IN LIVER CANCER | 22 | 19 | All SZGR 2.0 genes in this pathway |
| SUZUKI AMPLIFIED IN ORAL CANCER | 16 | 12 | All SZGR 2.0 genes in this pathway |
| MYLLYKANGAS AMPLIFICATION HOT SPOT 29 | 11 | 7 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS LUMINAL | 326 | 213 | All SZGR 2.0 genes in this pathway |
| HONRADO BREAST CANCER BRCA1 VS BRCA2 | 18 | 12 | All SZGR 2.0 genes in this pathway |
| SNIJDERS AMPLIFIED IN HEAD AND NECK TUMORS | 37 | 27 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| SIMBULAN UV RESPONSE NORMAL DN | 33 | 27 | All SZGR 2.0 genes in this pathway |
| SIMBULAN UV RESPONSE IMMORTALIZED DN | 31 | 26 | All SZGR 2.0 genes in this pathway |
| HASINA NOL7 TARGETS DN | 13 | 9 | All SZGR 2.0 genes in this pathway |
| LIU NASOPHARYNGEAL CARCINOMA | 70 | 38 | All SZGR 2.0 genes in this pathway |
| LOCKWOOD AMPLIFIED IN LUNG CANCER | 214 | 139 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR UP | 176 | 115 | All SZGR 2.0 genes in this pathway |
| COLIN PILOCYTIC ASTROCYTOMA VS GLIOBLASTOMA DN | 28 | 21 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 480 HELA | 164 | 118 | All SZGR 2.0 genes in this pathway |
| JEON SMAD6 TARGETS DN | 19 | 14 | All SZGR 2.0 genes in this pathway |
| SUNG METASTASIS STROMA DN | 54 | 34 | All SZGR 2.0 genes in this pathway |
| TCGA GLIOBLASTOMA COPY NUMBER UP | 75 | 36 | All SZGR 2.0 genes in this pathway |
| DING LUNG CANCER MUTATED SIGNIFICANTLY | 26 | 22 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 1 UP | 140 | 85 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER CIPROFIBRATE DN | 66 | 43 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC TGFA DN | 65 | 44 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER ACOX1 DN | 65 | 39 | All SZGR 2.0 genes in this pathway |
| SMITH TERT TARGETS UP | 145 | 91 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC E2F1 DN | 64 | 38 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC DN | 63 | 40 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER E2F1 DN | 64 | 42 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS UP | 214 | 133 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS MATURE CELL | 293 | 160 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS DN | 371 | 218 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN | 298 | 200 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE DN | 123 | 76 | All SZGR 2.0 genes in this pathway |
| YIH RESPONSE TO ARSENITE C4 | 18 | 12 | All SZGR 2.0 genes in this pathway |
| CHIBA RESPONSE TO TSA DN | 23 | 14 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN DN | 271 | 175 | All SZGR 2.0 genes in this pathway |
| SU PLACENTA | 30 | 18 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY 4NQO OR GAMMA RADIATION | 15 | 13 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE UP | 226 | 164 | All SZGR 2.0 genes in this pathway |
| KONDO EZH2 TARGETS | 245 | 148 | All SZGR 2.0 genes in this pathway |
| ZHANG GATA6 TARGETS DN | 64 | 46 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| DE YY1 TARGETS DN | 92 | 64 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR DN | 191 | 123 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BRAIN UP | 39 | 20 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE DN | 315 | 197 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL UP | 260 | 174 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| HUANG DASATINIB RESISTANCE UP | 81 | 53 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| SEKI INFLAMMATORY RESPONSE LPS UP | 77 | 56 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| RUIZ TNC TARGETS DN | 142 | 79 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE DN | 209 | 137 | All SZGR 2.0 genes in this pathway |
| HINATA NFKB TARGETS KERATINOCYTE UP | 91 | 63 | All SZGR 2.0 genes in this pathway |
| HINATA NFKB TARGETS FIBROBLAST UP | 84 | 60 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP | 395 | 249 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| DING LUNG CANCER MUTATED RECURRENTLY | 6 | 5 | All SZGR 2.0 genes in this pathway |
| TCGA GLIOBLASTOMA MUTATED | 8 | 8 | All SZGR 2.0 genes in this pathway |
| GYORFFY DOXORUBICIN RESISTANCE | 56 | 34 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S3 | 266 | 180 | All SZGR 2.0 genes in this pathway |
| NIELSEN SCHWANNOMA DN | 18 | 9 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA CLASSICAL | 162 | 122 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS LOW SERUM | 100 | 51 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS DN | 414 | 237 | All SZGR 2.0 genes in this pathway |
| OHGUCHI LIVER HNF4A TARGETS DN | 149 | 85 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS DN | 229 | 135 | All SZGR 2.0 genes in this pathway |
| ZHU SKIL TARGETS UP | 20 | 14 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| SERVITJA LIVER HNF1A TARGETS DN | 157 | 105 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 3 | 20 | 9 | All SZGR 2.0 genes in this pathway |
| PEDERSEN TARGETS OF 611CTF ISOFORM OF ERBB2 | 76 | 45 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| LIU IL13 PRIMING MODEL | 15 | 11 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE VIA P38 PARTIAL | 160 | 106 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38 | 337 | 236 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY LUMINAL MATURE DN | 99 | 74 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER CLUSTER 3 | 16 | 12 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-128 | 200 | 206 | 1A | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-27 | 200 | 207 | 1A,m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.