| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL UP
| 285 | 181 | All SZGR 2.0 genes in this pathway |
| KORKOLA TERATOMA UP
| 16 | 11 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP
| 783 | 507 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE UP
| 351 | 230 | All SZGR 2.0 genes in this pathway |
| DAVICIONI TARGETS OF PAX FOXO1 FUSIONS UP
| 255 | 177 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL DN
| 455 | 304 | All SZGR 2.0 genes in this pathway |
| LAIHO COLORECTAL CANCER SERRATED UP
| 112 | 71 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS TURQUOISE UP
| 79 | 50 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS GREY UP
| 18 | 10 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 8D DN
| 205 | 127 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 10D DN
| 142 | 90 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY DN
| 362 | 238 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC UP
| 185 | 126 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION ERYTHROCYTE UP
| 157 | 104 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION SUSTAINDED IN ERYTHROCYTE UP
| 44 | 30 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP
| 501 | 327 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR UP
| 164 | 122 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL UP
| 316 | 190 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML QUIESCENT VS CML DIVIDING UP
| 23 | 13 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML QUIESCENT VS NORMAL QUIESCENT DN
| 47 | 32 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML DIVIDING VS NORMAL QUIESCENT DN
| 95 | 57 | All SZGR 2.0 genes in this pathway |
| GRAHAM NORMAL QUIESCENT VS NORMAL DIVIDING UP
| 66 | 47 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN
| 805 | 505 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN
| 508 | 354 | All SZGR 2.0 genes in this pathway |
| KOINUMA COLON CANCER MSI DN
| 8 | 5 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN
| 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| CAIRO PML TARGETS BOUND BY MYC UP
| 23 | 17 | All SZGR 2.0 genes in this pathway |
| WANG BARRETTS ESOPHAGUS AND ESOPHAGUS CANCER DN
| 37 | 22 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP
| 584 | 356 | All SZGR 2.0 genes in this pathway |
| ROVERSI GLIOMA LOH REGIONS
| 44 | 30 | All SZGR 2.0 genes in this pathway |
| KORKOLA TERATOMA
| 39 | 25 | All SZGR 2.0 genes in this pathway |
| SIMBULAN UV RESPONSE NORMAL DN
| 33 | 27 | All SZGR 2.0 genes in this pathway |
| SIMBULAN UV RESPONSE IMMORTALIZED DN
| 31 | 26 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO PACLITAXEL VIA MAPK8 UP
| 14 | 9 | All SZGR 2.0 genes in this pathway |
| KHETCHOUMIAN TRIM24 TARGETS UP
| 47 | 38 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX
| 795 | 478 | All SZGR 2.0 genes in this pathway |
| CAFFAREL RESPONSE TO THC UP
| 33 | 20 | All SZGR 2.0 genes in this pathway |
| AMIT SERUM RESPONSE 240 MCF10A
| 57 | 36 | All SZGR 2.0 genes in this pathway |
| GEORGES CELL CYCLE MIR192 TARGETS
| 62 | 46 | All SZGR 2.0 genes in this pathway |
| LIN SILENCED BY TUMOR MICROENVIRONMENT
| 108 | 73 | All SZGR 2.0 genes in this pathway |
| MORI PRE BI LYMPHOCYTE UP
| 80 | 54 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL CIS
| 128 | 77 | All SZGR 2.0 genes in this pathway |
| BECKER TAMOXIFEN RESISTANCE DN
| 52 | 37 | All SZGR 2.0 genes in this pathway |
| SASAKI ADULT T CELL LEUKEMIA
| 176 | 122 | All SZGR 2.0 genes in this pathway |
| LENAOUR DENDRITIC CELL MATURATION DN
| 128 | 90 | All SZGR 2.0 genes in this pathway |
| GREENBAUM E2A TARGETS DN
| 21 | 11 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED DN
| 246 | 180 | All SZGR 2.0 genes in this pathway |
| IGLESIAS E2F TARGETS UP
| 151 | 103 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN
| 408 | 274 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 10HR DN
| 56 | 37 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C2
| 54 | 39 | All SZGR 2.0 genes in this pathway |
| JI RESPONSE TO FSH DN
| 58 | 43 | All SZGR 2.0 genes in this pathway |
| KRASNOSELSKAYA ILF3 TARGETS UP
| 38 | 24 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL
| 254 | 164 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP
| 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 8
| 86 | 57 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C1
| 72 | 45 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D8
| 40 | 29 | All SZGR 2.0 genes in this pathway |
| BILD MYC ONCOGENIC SIGNATURE
| 206 | 117 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 5
| 482 | 296 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P7
| 90 | 52 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED
| 1229 | 713 | All SZGR 2.0 genes in this pathway |
| RIGGINS TAMOXIFEN RESISTANCE DN
| 220 | 147 | All SZGR 2.0 genes in this pathway |
| KONDO EZH2 TARGETS
| 245 | 148 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP
| 578 | 341 | All SZGR 2.0 genes in this pathway |
| DE YY1 TARGETS UP
| 20 | 14 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR DN
| 354 | 216 | All SZGR 2.0 genes in this pathway |
| BASSO HAIRY CELL LEUKEMIA DN
| 80 | 66 | All SZGR 2.0 genes in this pathway |
| ZHENG GLIOBLASTOMA PLASTICITY DN
| 58 | 39 | All SZGR 2.0 genes in this pathway |
| LABBE WNT3A TARGETS DN
| 97 | 53 | All SZGR 2.0 genes in this pathway |
| LABBE TARGETS OF TGFB1 AND WNT3A DN
| 108 | 68 | All SZGR 2.0 genes in this pathway |
| ALONSO METASTASIS EMT UP
| 36 | 24 | All SZGR 2.0 genes in this pathway |
| ALONSO METASTASIS NEURAL UP
| 18 | 13 | All SZGR 2.0 genes in this pathway |
| ALONSO METASTASIS UP
| 198 | 128 | All SZGR 2.0 genes in this pathway |
| CROMER TUMORIGENESIS DN
| 51 | 29 | All SZGR 2.0 genes in this pathway |
| FLOTHO PEDIATRIC ALL THERAPY RESPONSE UP
| 53 | 33 | All SZGR 2.0 genes in this pathway |
| GOLDRATH ANTIGEN RESPONSE
| 346 | 192 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN
| 366 | 257 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 DN
| 315 | 201 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP
| 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP
| 408 | 276 | All SZGR 2.0 genes in this pathway |
| HOFFMANN PRE BI TO LARGE PRE BII LYMPHOCYTE DN
| 75 | 61 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP
| 395 | 249 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION
| 422 | 277 | All SZGR 2.0 genes in this pathway |
| GYORFFY DOXORUBICIN RESISTANCE
| 56 | 34 | All SZGR 2.0 genes in this pathway |
| DANG MYC TARGETS UP
| 143 | 100 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC
| 1103 | 714 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE
| 721 | 492 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3
| 718 | 401 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC ICP WITH H3K4ME3
| 445 | 257 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 9
| 76 | 45 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G2
| 182 | 102 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER UP
| 307 | 182 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT
| 572 | 352 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS UP
| 387 | 225 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS UP
| 217 | 131 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS GROUP2
| 60 | 38 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 DN
| 336 | 211 | All SZGR 2.0 genes in this pathway |
| ZHU SKIL TARGETS DN
| 9 | 5 | All SZGR 2.0 genes in this pathway |
| DELPUECH FOXO3 TARGETS UP
| 68 | 49 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP
| 682 | 433 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3
| 824 | 528 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP
| 570 | 339 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 4
| 110 | 66 | All SZGR 2.0 genes in this pathway |
| DELACROIX RARG BOUND MEF
| 367 | 231 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR TARGETS DN
| 24 | 16 | All SZGR 2.0 genes in this pathway |
| ALFANO MYC TARGETS
| 239 | 156 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN
| 784 | 464 | All SZGR 2.0 genes in this pathway |
| PEDRIOLI MIR31 TARGETS DN
| 418 | 245 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38
| 337 | 236 | All SZGR 2.0 genes in this pathway |
| PECE MAMMARY STEM CELL DN
| 146 | 88 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 3 TRANSIENTLY INDUCED BY EGF
| 222 | 159 | All SZGR 2.0 genes in this pathway |
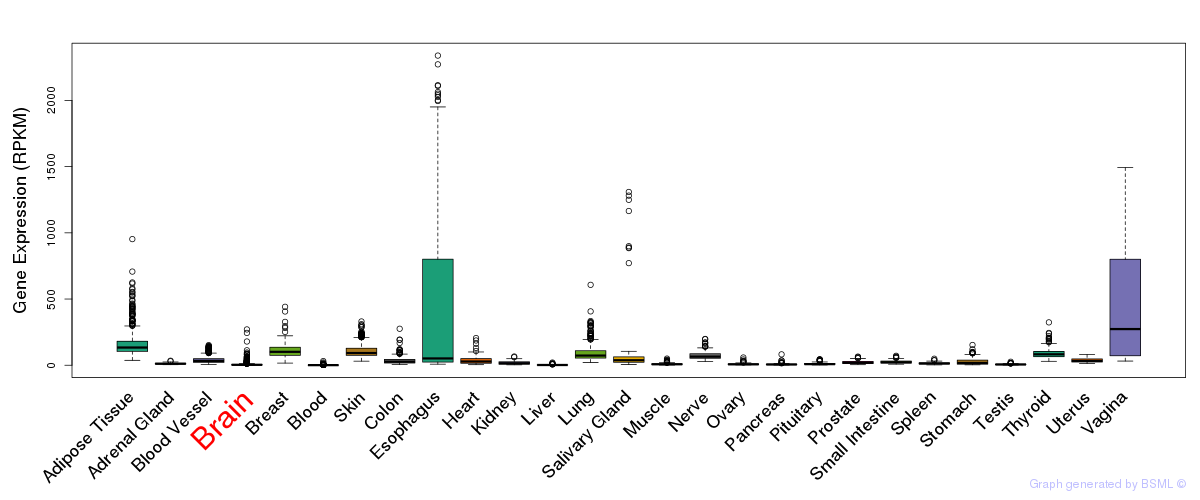
 Differentially methylated gene
Differentially methylated gene eQTL annotation
eQTL annotation