Gene Page: FOXC1
Summary ?
| GeneID | 2296 |
| Symbol | FOXC1 |
| Synonyms | ARA|FKHL7|FREAC-3|FREAC3|IGDA|IHG1|IRID1|RIEG3 |
| Description | forkhead box C1 |
| Reference | MIM:601090|HGNC:HGNC:3800|Ensembl:ENSG00000054598|HPRD:03054|Vega:OTTHUMG00000016182 |
| Gene type | protein-coding |
| Map location | 6p25 |
| Pascal p-value | 0.307 |
| Fetal beta | 1.557 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0159 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg23867624 | 6 | 1610197 | FOXC1 | 2.07E-8 | -0.009 | 7.09E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs12530461 | chr6 | 614352 | FOXC1 | 2296 | 0.04 | cis | ||
| rs1320385 | chr1 | 54064468 | FOXC1 | 2296 | 1.021E-4 | trans | ||
| rs12036675 | chr1 | 54066317 | FOXC1 | 2296 | 1.021E-4 | trans | ||
| rs12062528 | chr1 | 54070830 | FOXC1 | 2296 | 2.137E-4 | trans | ||
| rs12561793 | chr1 | 220647005 | FOXC1 | 2296 | 0.16 | trans | ||
| rs4522651 | chr2 | 34048 | FOXC1 | 2296 | 0 | trans | ||
| rs17077856 | chr3 | 45504326 | FOXC1 | 2296 | 0.12 | trans | ||
| rs2922180 | chr3 | 125280472 | FOXC1 | 2296 | 2.337E-6 | trans | ||
| rs2976809 | chr3 | 125451738 | FOXC1 | 2296 | 0.03 | trans | ||
| rs865499 | chr3 | 165446956 | FOXC1 | 2296 | 0.17 | trans | ||
| rs1216383 | chr4 | 129503677 | FOXC1 | 2296 | 0.19 | trans | ||
| rs7448349 | 0 | FOXC1 | 2296 | 3.582E-5 | trans | |||
| snp_a-2229839 | 0 | FOXC1 | 2296 | 0 | trans | |||
| rs248049 | chr5 | 126489320 | FOXC1 | 2296 | 0.13 | trans | ||
| rs816736 | chr5 | 154271947 | FOXC1 | 2296 | 2.953E-5 | trans | ||
| rs6934246 | chr6 | 35154494 | FOXC1 | 2296 | 0.02 | trans | ||
| rs4711782 | chr6 | 44363229 | FOXC1 | 2296 | 0.1 | trans | ||
| rs6455226 | chr6 | 67930295 | FOXC1 | 2296 | 0.2 | trans | ||
| rs12056282 | chr7 | 18882202 | FOXC1 | 2296 | 0.07 | trans | ||
| rs2739708 | chr8 | 17992280 | FOXC1 | 2296 | 0 | trans | ||
| rs714131 | chr8 | 17992832 | FOXC1 | 2296 | 0.01 | trans | ||
| rs10955813 | chr8 | 118415080 | FOXC1 | 2296 | 0.07 | trans | ||
| rs10969120 | chr9 | 29400711 | FOXC1 | 2296 | 0.02 | trans | ||
| rs4743989 | chr9 | 97859191 | FOXC1 | 2296 | 0.15 | trans | ||
| rs651134 | chr10 | 27549960 | FOXC1 | 2296 | 0.03 | trans | ||
| rs11006556 | chr10 | 61287609 | FOXC1 | 2296 | 0.15 | trans | ||
| rs1159612 | chr10 | 85329057 | FOXC1 | 2296 | 0 | trans | ||
| rs10886877 | chr10 | 85425690 | FOXC1 | 2296 | 0.16 | trans | ||
| rs7071168 | chr10 | 95829674 | FOXC1 | 2296 | 0.01 | trans | ||
| rs12572674 | chr10 | 107733982 | FOXC1 | 2296 | 0.07 | trans | ||
| rs4612791 | chr11 | 40768423 | FOXC1 | 2296 | 0.05 | trans | ||
| rs481843 | chr11 | 116525866 | FOXC1 | 2296 | 0.02 | trans | ||
| rs2909003 | chr12 | 12208768 | FOXC1 | 2296 | 0.04 | trans | ||
| rs11049036 | chr12 | 27650292 | FOXC1 | 2296 | 0.04 | trans | ||
| rs7303819 | chr12 | 33990776 | FOXC1 | 2296 | 0.1 | trans | ||
| rs10879027 | chr12 | 70237156 | FOXC1 | 2296 | 0.12 | trans | ||
| rs11115813 | chr12 | 83808239 | FOXC1 | 2296 | 2.957E-4 | trans | ||
| rs17073665 | chr13 | 81541961 | FOXC1 | 2296 | 0.01 | trans | ||
| rs1457420 | chr14 | 66958675 | FOXC1 | 2296 | 0.1 | trans | ||
| rs1872761 | chr15 | 40003815 | FOXC1 | 2296 | 0.18 | trans | ||
| rs190183 | chr15 | 97546708 | FOXC1 | 2296 | 0.15 | trans | ||
| rs17137819 | chr16 | 5372534 | FOXC1 | 2296 | 0.02 | trans | ||
| rs16958302 | chr16 | 57647432 | FOXC1 | 2296 | 0.01 | trans | ||
| rs9959804 | chr18 | 20264142 | FOXC1 | 2296 | 0 | trans | ||
| rs17066440 | chr18 | 57648274 | FOXC1 | 2296 | 0.03 | trans | ||
| rs17001072 | chr19 | 11099774 | FOXC1 | 2296 | 0.04 | trans | ||
| rs6095741 | chr20 | 48666589 | FOXC1 | 2296 | 0 | trans | ||
| rs7265852 | chr20 | 48823717 | FOXC1 | 2296 | 0.03 | trans | ||
| rs5991662 | chrX | 43335796 | FOXC1 | 2296 | 0.11 | trans | ||
| rs1545676 | chrX | 93530472 | FOXC1 | 2296 | 2.545E-4 | trans | ||
| rs242163 | chrX | 132311662 | FOXC1 | 2296 | 0.04 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
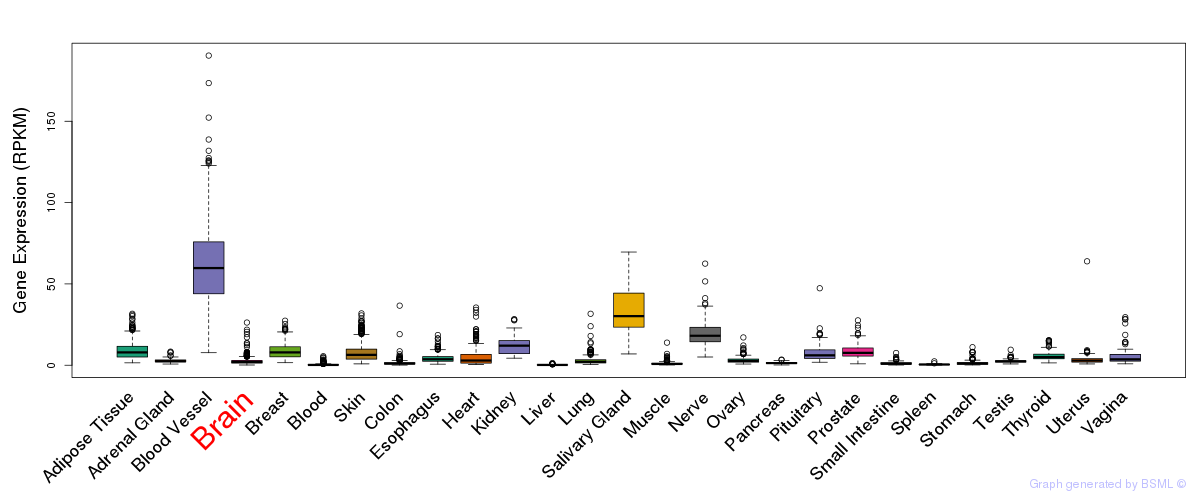
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PLXNA1 | 0.88 | 0.87 |
| ZNF778 | 0.86 | 0.82 |
| NAV1 | 0.86 | 0.85 |
| CELSR3 | 0.86 | 0.81 |
| CACNA1E | 0.85 | 0.81 |
| KIAA1549 | 0.85 | 0.82 |
| DLG5 | 0.85 | 0.85 |
| ZNF629 | 0.85 | 0.84 |
| ANKS1A | 0.85 | 0.83 |
| AMMECR1L | 0.84 | 0.84 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.57 | -0.66 |
| COPZ2 | -0.55 | -0.66 |
| AF347015.31 | -0.55 | -0.72 |
| AF347015.27 | -0.54 | -0.70 |
| S100B | -0.54 | -0.68 |
| MT-CO2 | -0.54 | -0.71 |
| HLA-F | -0.54 | -0.57 |
| AIFM3 | -0.54 | -0.58 |
| TSC22D4 | -0.53 | -0.62 |
| FXYD1 | -0.53 | -0.69 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003705 | RNA polymerase II transcription factor activity, enhancer binding | IEA | - | |
| GO:0008301 | DNA bending activity | IDA | 7957066 | |
| GO:0008134 | transcription factor binding | IPI | 15684392 | |
| GO:0016563 | transcription activator activity | NAS | 8499623 | |
| GO:0043565 | sequence-specific DNA binding | IDA | 7957066 |15277473 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007420 | brain development | IEA | Brain (GO term level: 7) | - |
| GO:0001568 | blood vessel development | IEA | - | |
| GO:0001501 | skeletal system development | IEA | - | |
| GO:0001503 | ossification | IEA | - | |
| GO:0001657 | ureteric bud development | IEA | - | |
| GO:0001701 | in utero embryonic development | IEA | - | |
| GO:0001541 | ovarian follicle development | IEA | - | |
| GO:0003007 | heart morphogenesis | IEA | - | |
| GO:0001822 | kidney development | IEA | - | |
| GO:0001945 | lymph vessel development | IEA | - | |
| GO:0001974 | blood vessel remodeling | IEA | - | |
| GO:0055010 | ventricular cardiac muscle morphogenesis | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0007219 | Notch signaling pathway | IEA | - | |
| GO:0008354 | germ cell migration | IEA | - | |
| GO:0014032 | neural crest cell development | IEA | - | |
| GO:0048010 | vascular endothelial growth factor receptor signaling pathway | IEA | - | |
| GO:0006916 | anti-apoptosis | IEA | - | |
| GO:0030203 | glycosaminoglycan metabolic process | IEA | - | |
| GO:0042475 | odontogenesis of dentine-containing tooth | IMP | 12614756 | |
| GO:0030199 | collagen fibril organization | IEA | - | |
| GO:0043010 | camera-type eye development | IEA | - | |
| GO:0035050 | embryonic heart tube development | IEA | - | |
| GO:0032808 | lacrimal gland development | IEA | - | |
| GO:0048341 | paraxial mesoderm formation | IEA | - | |
| GO:0045930 | negative regulation of mitotic cell cycle | IDA | 12408963 | |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | IEA | - | |
| GO:0050880 | regulation of blood vessel size | IEA | - | |
| GO:0046620 | regulation of organ growth | IEA | - | |
| GO:0048844 | artery morphogenesis | IEA | - | |
| GO:0048762 | mesenchymal cell differentiation | IEA | - | |
| GO:0060038 | cardiac muscle cell proliferation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IC | 7957066 | |
| GO:0005634 | nucleus | IDA | 15277473 |16449236 | |
| GO:0005720 | nuclear heterochromatin | IDA | 15684392 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| CHEMNITZ RESPONSE TO PROSTAGLANDIN E2 DN | 391 | 222 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| DOANE BREAST CANCER CLASSES DN | 34 | 26 | All SZGR 2.0 genes in this pathway |
| DOANE BREAST CANCER ESR1 DN | 48 | 28 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS GREY DN | 74 | 44 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 8D UP | 157 | 91 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION DN | 329 | 219 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS UP | 214 | 155 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| TOMIDA METASTASIS DN | 18 | 11 | All SZGR 2.0 genes in this pathway |
| WONG ENDMETRIUM CANCER UP | 25 | 17 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION 6HR | 40 | 23 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS BASAL | 330 | 217 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER BASAL VS LULMINAL | 330 | 215 | All SZGR 2.0 genes in this pathway |
| LIEN BREAST CARCINOMA METAPLASTIC VS DUCTAL UP | 83 | 51 | All SZGR 2.0 genes in this pathway |
| DAWSON METHYLATED IN LYMPHOMA TCL1 | 59 | 45 | All SZGR 2.0 genes in this pathway |
| YANG BREAST CANCER ESR1 LASER DN | 50 | 38 | All SZGR 2.0 genes in this pathway |
| LIU NASOPHARYNGEAL CARCINOMA | 70 | 38 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES CORE NINE CORRELATED | 100 | 68 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 60 HELA | 46 | 32 | All SZGR 2.0 genes in this pathway |
| OKUMURA INFLAMMATORY RESPONSE LPS | 183 | 115 | All SZGR 2.0 genes in this pathway |
| ALCALAY AML BY NPM1 LOCALIZATION UP | 140 | 83 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED UP | 183 | 111 | All SZGR 2.0 genes in this pathway |
| ZHENG RESPONSE TO ARSENITE DN | 18 | 15 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK UP | 244 | 151 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS UP | 388 | 234 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE DN | 315 | 197 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| BRUECKNER TARGETS OF MIRLET7A3 UP | 111 | 69 | All SZGR 2.0 genes in this pathway |
| GU PDEF TARGETS DN | 39 | 20 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER ESR1 DN | 240 | 153 | All SZGR 2.0 genes in this pathway |
| COLINA TARGETS OF 4EBP1 AND 4EBP2 | 356 | 214 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 6 | 33 | 22 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED | 351 | 238 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS | 535 | 325 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 4HR DN | 254 | 158 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS NOT VIA AKT1 UP | 211 | 131 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 AND SATB1 UP | 87 | 52 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN | 448 | 282 | All SZGR 2.0 genes in this pathway |
| SCHMIDT POR TARGETS IN LIMB BUD UP | 26 | 21 | All SZGR 2.0 genes in this pathway |
| KATSANOU ELAVL1 TARGETS UP | 169 | 105 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR DN | 244 | 157 | All SZGR 2.0 genes in this pathway |
| WANG MLL TARGETS | 289 | 188 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE VIA P38 COMPLETE | 227 | 151 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-103/107 | 1755 | 1761 | 1A | hsa-miR-103brain | AGCAGCAUUGUACAGGGCUAUGA |
| hsa-miR-107brain | AGCAGCAUUGUACAGGGCUAUCA | ||||
| miR-133 | 538 | 544 | 1A | hsa-miR-133a | UUGGUCCCCUUCAACCAGCUGU |
| hsa-miR-133b | UUGGUCCCCUUCAACCAGCUA | ||||
| miR-137 | 1398 | 1405 | 1A,m8 | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| miR-138 | 160 | 167 | 1A,m8 | hsa-miR-138brain | AGCUGGUGUUGUGAAUC |
| hsa-miR-138brain | AGCUGGUGUUGUGAAUC | ||||
| miR-204/211 | 619 | 626 | 1A,m8 | hsa-miR-204brain | UUCCCUUUGUCAUCCUAUGCCU |
| hsa-miR-211 | UUCCCUUUGUCAUCCUUCGCCU | ||||
| miR-24 | 1166 | 1172 | 1A | hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG |
| miR-28 | 1681 | 1687 | 1A | hsa-miR-28brain | AAGGAGCUCACAGUCUAUUGAG |
| miR-320 | 746 | 752 | 1A | hsa-miR-320 | AAAAGCUGGGUUGAGAGGGCGAA |
| miR-330 | 928 | 935 | 1A,m8 | hsa-miR-330brain | GCAAAGCACACGGCCUGCAGAGA |
| miR-374 | 612 | 618 | 1A | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| miR-495 | 682 | 688 | m8 | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
| hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU | ||||
| miR-496 | 488 | 494 | 1A | hsa-miR-496 | AUUACAUGGCCAAUCUC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.