| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN
| 634 | 384 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA DN
| 349 | 157 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN
| 493 | 298 | All SZGR 2.0 genes in this pathway |
| CHEMNITZ RESPONSE TO PROSTAGLANDIN E2 DN
| 391 | 222 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 DN
| 175 | 82 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER BASAL VS MESENCHYMAL UP
| 121 | 45 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS DN
| 384 | 230 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY UP
| 430 | 232 | All SZGR 2.0 genes in this pathway |
| SABATES COLORECTAL ADENOMA UP
| 141 | 75 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP
| 1821 | 933 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN
| 781 | 465 | All SZGR 2.0 genes in this pathway |
| PROVENZANI METASTASIS DN
| 136 | 94 | All SZGR 2.0 genes in this pathway |
| LANDIS BREAST CANCER PROGRESSION UP
| 44 | 27 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP
| 443 | 294 | All SZGR 2.0 genes in this pathway |
| CASTELLANO NRAS TARGETS UP
| 68 | 41 | All SZGR 2.0 genes in this pathway |
| LANDIS ERBB2 BREAST PRENEOPLASTIC UP
| 20 | 10 | All SZGR 2.0 genes in this pathway |
| LANDIS ERBB2 BREAST TUMORS 324 UP
| 150 | 93 | All SZGR 2.0 genes in this pathway |
| MISSIAGLIA REGULATED BY METHYLATION UP
| 126 | 78 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 3 4WK UP
| 214 | 144 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK UP
| 271 | 175 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL TGFB1 TARGETS DN
| 62 | 44 | All SZGR 2.0 genes in this pathway |
| WONG ENDMETRIUM CANCER UP
| 25 | 17 | All SZGR 2.0 genes in this pathway |
| MUELLER METHYLATED IN GLIOBLASTOMA
| 40 | 22 | All SZGR 2.0 genes in this pathway |
| HUMMERICH SKIN CANCER PROGRESSION UP
| 88 | 58 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 2
| 127 | 92 | All SZGR 2.0 genes in this pathway |
| RASHI NFKB1 TARGETS
| 19 | 18 | All SZGR 2.0 genes in this pathway |
| LI AMPLIFIED IN LUNG CANCER
| 178 | 108 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER
| 747 | 453 | All SZGR 2.0 genes in this pathway |
| KORKOLA TERATOMA
| 39 | 25 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP
| 358 | 245 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM3
| 70 | 37 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN
| 848 | 527 | All SZGR 2.0 genes in this pathway |
| GOUYER TATI TARGETS UP
| 10 | 7 | All SZGR 2.0 genes in this pathway |
| LIN SILENCED BY TUMOR MICROENVIRONMENT
| 108 | 73 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 DN
| 464 | 276 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 1 DN
| 169 | 102 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 3 DN
| 59 | 32 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC UP
| 54 | 30 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC TGFA UP
| 61 | 40 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP
| 398 | 262 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK UP
| 87 | 58 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER ACOX1 UP
| 64 | 40 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC E2F1 UP
| 56 | 34 | All SZGR 2.0 genes in this pathway |
| CHEOK RESPONSE TO MERCAPTOPURINE AND LD MTX DN
| 21 | 11 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER CIPROFIBRATE UP
| 60 | 42 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER DENA UP
| 60 | 40 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER E2F1 UP
| 62 | 35 | All SZGR 2.0 genes in this pathway |
| YAO HOXA10 TARGETS VIA PROGESTERONE UP
| 79 | 58 | All SZGR 2.0 genes in this pathway |
| BROWN MYELOID CELL DEVELOPMENT UP
| 165 | 100 | All SZGR 2.0 genes in this pathway |
| LEI MYB TARGETS
| 318 | 215 | All SZGR 2.0 genes in this pathway |
| HESS TARGETS OF HOXA9 AND MEIS1 DN
| 77 | 48 | All SZGR 2.0 genes in this pathway |
| ABBUD LIF SIGNALING 1 UP
| 46 | 29 | All SZGR 2.0 genes in this pathway |
| GERY CEBP TARGETS
| 126 | 90 | All SZGR 2.0 genes in this pathway |
| LIAN NEUTROPHIL GRANULE CONSTITUENTS
| 25 | 13 | All SZGR 2.0 genes in this pathway |
| RADMACHER AML PROGNOSIS
| 78 | 52 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED DN
| 246 | 180 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA DN
| 41 | 27 | All SZGR 2.0 genes in this pathway |
| LIANG SILENCED BY METHYLATION 2
| 53 | 34 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR UP
| 240 | 152 | All SZGR 2.0 genes in this pathway |
| MCDOWELL ACUTE LUNG INJURY UP
| 45 | 29 | All SZGR 2.0 genes in this pathway |
| CHANG IMMORTALIZED BY HPV31 DN
| 65 | 45 | All SZGR 2.0 genes in this pathway |
| PAL PRMT5 TARGETS DN
| 29 | 16 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS PEAK AT 24HR
| 43 | 30 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C1
| 72 | 45 | All SZGR 2.0 genes in this pathway |
| HAN JNK SINGALING DN
| 39 | 27 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 6
| 189 | 112 | All SZGR 2.0 genes in this pathway |
| YANG MUC2 TARGETS DUODENUM 6MO UP
| 10 | 5 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3
| 720 | 440 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE UP
| 156 | 92 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY UV
| 62 | 43 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE DN
| 195 | 135 | All SZGR 2.0 genes in this pathway |
| STEARMAN TUMOR FIELD EFFECT UP
| 36 | 22 | All SZGR 2.0 genes in this pathway |
| STEARMAN LUNG CANCER EARLY VS LATE DN
| 61 | 39 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP
| 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP
| 863 | 514 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN
| 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP
| 648 | 398 | All SZGR 2.0 genes in this pathway |
| LABBE TGFB1 TARGETS DN
| 108 | 64 | All SZGR 2.0 genes in this pathway |
| LABBE TARGETS OF TGFB1 AND WNT3A DN
| 108 | 68 | All SZGR 2.0 genes in this pathway |
| HUPER BREAST BASAL VS LUMINAL DN
| 59 | 44 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC
| 420 | 269 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C D DN
| 252 | 155 | All SZGR 2.0 genes in this pathway |
| LUDWICZEK TREATING IRON OVERLOAD
| 7 | 5 | All SZGR 2.0 genes in this pathway |
| MARTINELLI IMMATURE NEUTROPHIL UP
| 11 | 6 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP
| 1305 | 895 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP
| 491 | 316 | All SZGR 2.0 genes in this pathway |
| SWEET KRAS ONCOGENIC SIGNATURE
| 89 | 56 | All SZGR 2.0 genes in this pathway |
| COLINA TARGETS OF 4EBP1 AND 4EBP2
| 356 | 214 | All SZGR 2.0 genes in this pathway |
| VALK AML WITH CEBPA
| 37 | 27 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA PCA1 UP
| 101 | 66 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT DN
| 222 | 141 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 5
| 30 | 22 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 30MIN UP
| 56 | 38 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 4HR UP
| 55 | 41 | All SZGR 2.0 genes in this pathway |
| KAMIKUBO MYELOID CEBPA NETWORK
| 28 | 19 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS DN
| 242 | 146 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS DN
| 213 | 127 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN
| 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B
| 549 | 316 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP
| 745 | 475 | All SZGR 2.0 genes in this pathway |
| GUILLAUMOND KLF10 TARGETS UP
| 51 | 39 | All SZGR 2.0 genes in this pathway |
| BOSCO EPITHELIAL DIFFERENTIATION MODULE
| 69 | 31 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL UP
| 289 | 184 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY
| 1839 | 928 | All SZGR 2.0 genes in this pathway |
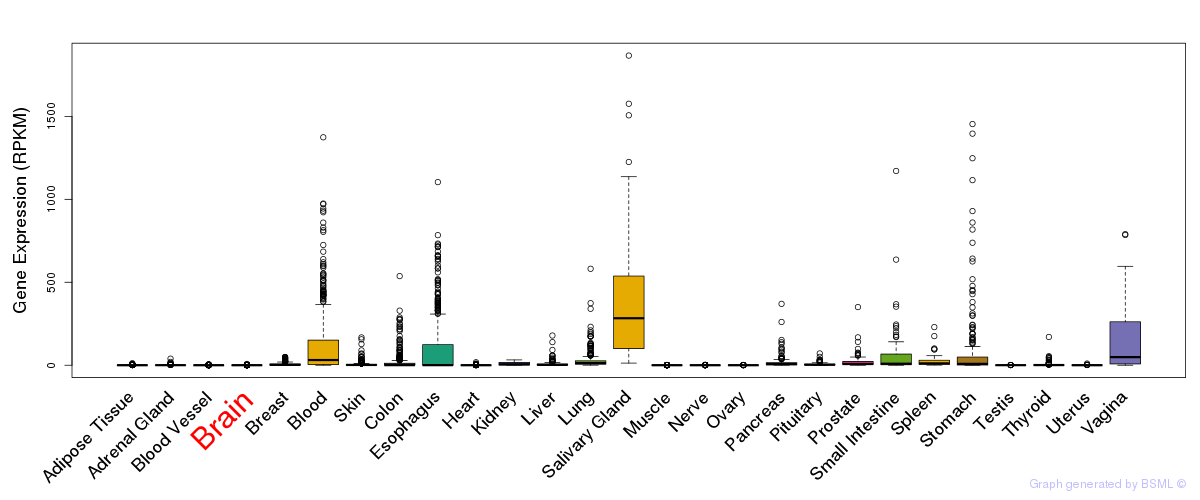
 eQTL annotation
eQTL annotation