Gene Page: ABLIM1
Summary ?
| GeneID | 3983 |
| Symbol | ABLIM1 |
| Synonyms | ABLIM|LIMAB1|LIMATIN|abLIM-1 |
| Description | actin binding LIM protein 1 |
| Reference | MIM:602330|HGNC:HGNC:78|Ensembl:ENSG00000099204|HPRD:03819|Vega:OTTHUMG00000019088 |
| Gene type | protein-coding |
| Map location | 10q25 |
| Pascal p-value | 0.122 |
| Sherlock p-value | 0.843 |
| Fetal beta | -0.71 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | STRUCTURAL PLASTICITY G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_TAP-PSD-95-CORE G2Cdb.humanNRC CompositeSet Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg12649038 | 10 | 116282534 | ABLIM1 | 2.72E-5 | 0.502 | 0.018 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16829545 | chr2 | 151977407 | ABLIM1 | 3983 | 7.857E-9 | trans | ||
| rs3845734 | chr2 | 171125572 | ABLIM1 | 3983 | 0 | trans | ||
| rs7584986 | chr2 | 184111432 | ABLIM1 | 3983 | 6.216E-6 | trans | ||
| rs2183142 | chr4 | 159232695 | ABLIM1 | 3983 | 0.13 | trans | ||
| rs9461864 | chr6 | 33481468 | ABLIM1 | 3983 | 0.15 | trans | ||
| rs3118341 | chr9 | 25185518 | ABLIM1 | 3983 | 0.06 | trans | ||
| rs16955618 | chr15 | 29937543 | ABLIM1 | 3983 | 2.44E-16 | trans | ||
| rs1041786 | chr21 | 22617710 | ABLIM1 | 3983 | 4.22E-4 | trans |
Section II. Transcriptome annotation
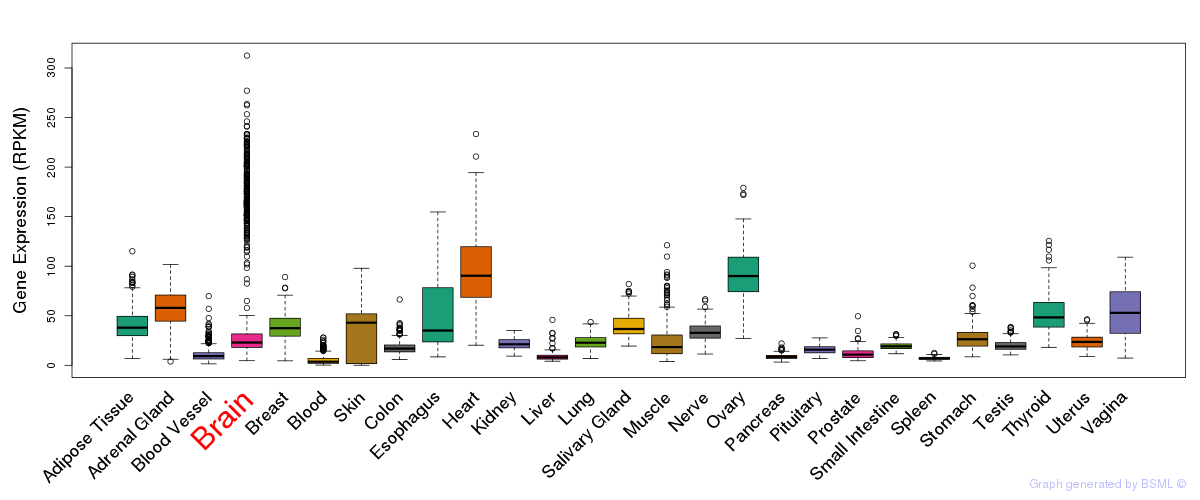
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FAM59A | 0.90 | 0.86 |
| CTTNBP2 | 0.90 | 0.85 |
| RP1-21O18.1 | 0.89 | 0.86 |
| LIMCH1 | 0.89 | 0.85 |
| OSBPL10 | 0.89 | 0.86 |
| GPD1L | 0.89 | 0.87 |
| TIAM2 | 0.89 | 0.88 |
| AFF3 | 0.89 | 0.88 |
| WASF1 | 0.88 | 0.86 |
| TBR1 | 0.88 | 0.86 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SERPINB6 | -0.67 | -0.73 |
| RHOC | -0.62 | -0.77 |
| HSD17B14 | -0.61 | -0.75 |
| BDH2 | -0.59 | -0.71 |
| FAH | -0.58 | -0.68 |
| S100A16 | -0.58 | -0.73 |
| TTYH1 | -0.57 | -0.64 |
| AF347015.31 | -0.57 | -0.72 |
| DBI | -0.57 | -0.76 |
| FXYD1 | -0.57 | -0.72 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BEGAIN | KIAA1446 | brain-enriched guanylate kinase-associated homolog (rat) | Two-hybrid | BioGRID | 16189514 |
| CALCOCO2 | MGC17318 | NDP52 | calcium binding and coiled-coil domain 2 | Two-hybrid | BioGRID | 16189514 |
| GOLGA2 | GM130 | MGC20672 | golgi autoantigen, golgin subfamily a, 2 | Two-hybrid | BioGRID | 16189514 |
| HOOK2 | FLJ26218 | HK2 | hook homolog 2 (Drosophila) | Two-hybrid | BioGRID | 16189514 |
| KCNJ12 | FLJ14167 | IRK2 | KCNJN1 | Kir2.2 | Kir2.2v | hIRK | hIRK1 | hkir2.2x | kcnj12x | potassium inwardly-rectifying channel, subfamily J, member 12 | - | HPRD,BioGRID | 15024025 |
| KRT15 | CK15 | K15 | K1CO | keratin 15 | Two-hybrid | BioGRID | 16189514 |
| LDOC1 | BCUR1 | Mar7 | Mart7 | leucine zipper, down-regulated in cancer 1 | Affinity Capture-Western Two-hybrid | BioGRID | 16189514 |
| SFN | YWHAS | stratifin | Affinity Capture-MS | BioGRID | 15778465 |
| TRAF2 | MGC:45012 | TRAP | TRAP3 | TNF receptor-associated factor 2 | Two-hybrid | BioGRID | 16189514 |
| TSC22D4 | THG-1 | THG1 | TSC22 domain family, member 4 | Two-hybrid | BioGRID | 16189514 |
| USHBP1 | AIEBP | FLJ38709 | FLJ90681 | MCC2 | Usher syndrome 1C binding protein 1 | Two-hybrid | BioGRID | 16189514 |
| VIM | FLJ36605 | vimentin | Two-hybrid | BioGRID | 16189514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG AXON GUIDANCE | 129 | 103 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME AXON GUIDANCE | 251 | 188 | All SZGR 2.0 genes in this pathway |
| REACTOME NETRIN1 SIGNALING | 41 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | 13 | 11 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA DN | 349 | 157 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL UP | 133 | 78 | All SZGR 2.0 genes in this pathway |
| JAEGER METASTASIS DN | 258 | 141 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 10D UP | 194 | 122 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS DN | 232 | 139 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN | 536 | 332 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION KRAS DN | 142 | 95 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 DN | 229 | 142 | All SZGR 2.0 genes in this pathway |
| VANHARANTA UTERINE FIBROID DN | 67 | 45 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS F UP | 185 | 119 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE AUGMENTED BY MYC | 108 | 74 | All SZGR 2.0 genes in this pathway |
| MATTIOLI MULTIPLE MYELOMA WITH 14Q32 TRANSLOCATIONS | 36 | 25 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| MORI PRE BI LYMPHOCYTE DN | 77 | 49 | All SZGR 2.0 genes in this pathway |
| MORI PLASMA CELL DN | 33 | 20 | All SZGR 2.0 genes in this pathway |
| LEE TARGETS OF PTCH1 AND SUFU DN | 83 | 69 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 DN | 464 | 276 | All SZGR 2.0 genes in this pathway |
| CHEOK RESPONSE TO HD MTX UP | 23 | 13 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA MF UP | 47 | 27 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED DN | 246 | 180 | All SZGR 2.0 genes in this pathway |
| KUMAR TARGETS OF MLL AF9 FUSION | 405 | 264 | All SZGR 2.0 genes in this pathway |
| BRACHAT RESPONSE TO CISPLATIN | 22 | 11 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL LONG TERM | 302 | 191 | All SZGR 2.0 genes in this pathway |
| RAMALHO STEMNESS DN | 74 | 55 | All SZGR 2.0 genes in this pathway |
| RAMASWAMY METASTASIS DN | 61 | 47 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BRACHAT RESPONSE TO METHOTREXATE UP | 26 | 16 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE DN | 261 | 183 | All SZGR 2.0 genes in this pathway |
| MCLACHLAN DENTAL CARIES DN | 245 | 144 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C5 | 46 | 36 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 24HR UP | 148 | 96 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE UP | 393 | 244 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS UP | 388 | 234 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| BRUECKNER TARGETS OF MIRLET7A3 UP | 111 | 69 | All SZGR 2.0 genes in this pathway |
| SEKI INFLAMMATORY RESPONSE LPS DN | 23 | 16 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SURVIVAL UP | 73 | 49 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS | 535 | 325 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 0 | 76 | 54 | All SZGR 2.0 genes in this pathway |
| ZEMBUTSU SENSITIVITY TO VINCRISTINE | 19 | 10 | All SZGR 2.0 genes in this pathway |
| NIELSEN LEIOMYOSARCOMA DN | 18 | 11 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO GONADOTROPHINS DN | 87 | 66 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO FORSKOLIN DN | 88 | 68 | All SZGR 2.0 genes in this pathway |
| WANG METASTASIS OF BREAST CANCER ESR1 DN | 30 | 18 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 1 UP | 125 | 61 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 2 UP | 54 | 31 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 3 UP | 170 | 97 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 4 UP | 112 | 64 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 6 UP | 140 | 81 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 7 UP | 118 | 68 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| KATSANOU ELAVL1 TARGETS UP | 169 | 105 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR DN | 244 | 157 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |