Gene Page: MAP2
Summary ?
| GeneID | 4133 |
| Symbol | MAP2 |
| Synonyms | MAP2A|MAP2B|MAP2C |
| Description | microtubule associated protein 2 |
| Reference | MIM:157130|HGNC:HGNC:6839|Ensembl:ENSG00000078018|HPRD:01140|Vega:OTTHUMG00000132962 |
| Gene type | protein-coding |
| Map location | 2q34-q35 |
| Pascal p-value | 0.032 |
| Fetal beta | 0.579 |
| eGene | Myers' cis & trans |
| Support | STRUCTURAL PLASTICITY G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_mGluR5 G2Cdb.humanNRC CompositeSet Darnell FMRP targets Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00916 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01016 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 1.2138 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs235427 | chr8 | 133815575 | MAP2 | 4133 | 0.17 | trans | ||
| rs7894597 | chr10 | 9384581 | MAP2 | 4133 | 0.03 | trans | ||
| rs1324669 | chr13 | 107872446 | MAP2 | 4133 | 0.16 | trans | ||
| rs17188417 | chr14 | 90517901 | MAP2 | 4133 | 0.2 | trans | ||
| rs6098023 | chr20 | 53113740 | MAP2 | 4133 | 0.17 | trans | ||
| rs17145698 | chrX | 40218345 | MAP2 | 4133 | 0.03 | trans |
Section II. Transcriptome annotation
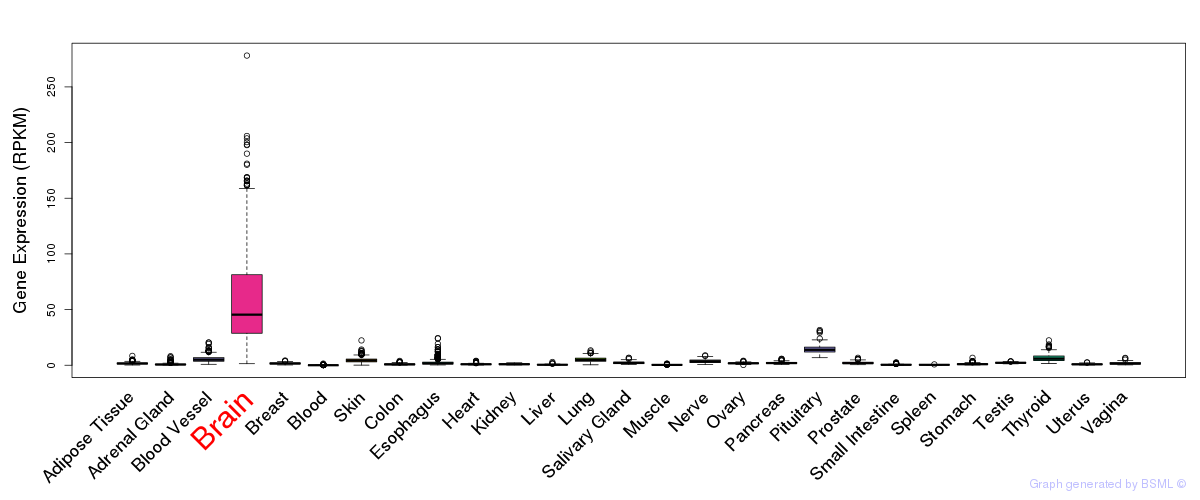
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003674 | molecular_function | ND | - | |
| GO:0005516 | calmodulin binding | IEA | - | |
| GO:0005198 | structural molecule activity | NAS | 9588626 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007026 | negative regulation of microtubule depolymerization | IEA | - | |
| GO:0008150 | biological_process | ND | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005856 | cytoskeleton | IEA | - | |
| GO:0005874 | microtubule | IEA | - | |
| GO:0005875 | microtubule associated complex | NAS | 7854050 | |
| GO:0005875 | microtubule associated complex | TAS | 9588626 | |
| GO:0005737 | cytoplasm | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| APOE | AD2 | LPG | MGC1571 | apoprotein | apolipoprotein E | - | HPRD | 8624078 |
| CPEB1 | CEBP | CPE-BP1 | CPEB | FLJ13203 | MGC34136 | MGC60106 | cytoplasmic polyadenylation element binding protein 1 | - | HPRD | 12629046 |
| FYN | MGC45350 | SLK | SYN | FYN oncogene related to SRC, FGR, YES | Affinity Capture-Western | BioGRID | 11546790 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | - | HPRD,BioGRID | 10781592 |11546790|11546790 |
| GRIN1 | NMDA1 | NMDAR1 | NR1 | glutamate receptor, ionotropic, N-methyl D-aspartate 1 | - | HPRD | 10862698 |
| GRIN2D | EB11 | NMDAR2D | glutamate receptor, ionotropic, N-methyl D-aspartate 2D | - | HPRD | 10862698 |
| MAP2 | DKFZp686I2148 | MAP2A | MAP2B | MAP2C | microtubule-associated protein 2 | - | HPRD,BioGRID | 10527895 |
| MARK4 | FLJ90097 | KIAA1860 | MARKL1 | Nbla00650 | MAP/microtubule affinity-regulating kinase 4 | - | HPRD,BioGRID | 14594945 |
| MYO7A | DFNA11 | DFNB2 | MYOVIIA | MYU7A | NSRD2 | USH1B | myosin VIIA | - | HPRD,BioGRID | 11171103 |
| NEFL | CMT1F | CMT2E | FLJ53642 | NF-L | NF68 | NFL | neurofilament, light polypeptide | Reconstituted Complex | BioGRID | 1902666 |
| PLEC1 | EBS1 | EBSO | HD1 | PCN | PLEC1b | PLTN | plectin 1, intermediate filament binding protein 500kDa | - | HPRD,BioGRID | 3027087 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | Reconstituted Complex | BioGRID | 10781592 |
| TTBK1 | BDTK | KIAA1855 | RP3-330M21.4 | tau tubulin kinase 1 | - | HPRD | 7556643 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA AKAPCENTROSOME PATHWAY | 15 | 14 | All SZGR 2.0 genes in this pathway |
| PID LKB1 PATHWAY | 47 | 37 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 DN | 175 | 82 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST DUCTAL CARCINOMA VS DUCTAL NORMAL DN | 198 | 110 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION DN | 329 | 219 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA DN | 77 | 52 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN UP | 239 | 157 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS NEUROEPITHELIUM DN | 164 | 111 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS | 539 | 324 | All SZGR 2.0 genes in this pathway |
| GUENTHER GROWTH SPHERICAL VS ADHERENT UP | 21 | 15 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN DN | 153 | 120 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 10HR UP | 101 | 69 | All SZGR 2.0 genes in this pathway |
| MAHAJAN RESPONSE TO IL1A UP | 81 | 52 | All SZGR 2.0 genes in this pathway |
| LEIN LOCALIZED TO PROXIMAL DENDRITES | 37 | 26 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE DN | 258 | 160 | All SZGR 2.0 genes in this pathway |
| MASSARWEH RESPONSE TO ESTRADIOL | 61 | 47 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN DN | 353 | 226 | All SZGR 2.0 genes in this pathway |
| HUANG FOXA2 TARGETS UP | 45 | 28 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| LABBE TARGETS OF TGFB1 AND WNT3A UP | 111 | 70 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| FONTAINE FOLLICULAR THYROID ADENOMA DN | 68 | 45 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA PR DN | 44 | 34 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA PRONEURAL | 177 | 132 | All SZGR 2.0 genes in this pathway |
| HOELZEL NF1 TARGETS UP | 139 | 93 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR DN | 505 | 328 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 DN | 336 | 211 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE EARLY LATE | 317 | 190 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS DN | 229 | 135 | All SZGR 2.0 genes in this pathway |
| TERAO AOX4 TARGETS SKIN UP | 38 | 27 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR DN | 244 | 157 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS UP | 279 | 155 | All SZGR 2.0 genes in this pathway |
| FOSTER KDM1A TARGETS UP | 266 | 142 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-129-5p | 3477 | 3483 | m8 | hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC |
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| miR-139 | 121 | 127 | m8 | hsa-miR-139brain | UCUACAGUGCACGUGUCU |
| miR-141/200a | 481 | 487 | 1A | hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG |
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| miR-17-5p/20/93.mr/106/519.d | 658 | 664 | m8 | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-186 | 82 | 89 | 1A,m8 | hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU |
| miR-194 | 3589 | 3595 | m8 | hsa-miR-194 | UGUAACAGCAACUCCAUGUGGA |
| hsa-miR-194 | UGUAACAGCAACUCCAUGUGGA | ||||
| miR-200bc/429 | 264 | 270 | 1A | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC | ||||
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-214 | 2922 | 2929 | 1A,m8 | hsa-miR-214brain | ACAGCAGGCACAGACAGGCAG |
| miR-23 | 893 | 899 | 1A | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-26 | 822 | 829 | 1A,m8 | hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC |
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
| miR-30-5p | 1115 | 1121 | 1A | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-335 | 43 | 49 | m8 | hsa-miR-335brain | UCAAGAGCAAUAACGAAAAAUGU |
| miR-338 | 3515 | 3521 | m8 | hsa-miR-338brain | UCCAGCAUCAGUGAUUUUGUUGA |
| miR-361 | 3572 | 3579 | 1A,m8 | hsa-miR-361brain | UUAUCAGAAUCUCCAGGGGUAC |
| miR-369-3p | 265 | 271 | m8 | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU | ||||
| miR-374 | 764 | 770 | m8 | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG | ||||
| miR-381 | 490 | 496 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-450 | 3477 | 3483 | 1A | hsa-miR-450 | UUUUUGCGAUGUGUUCCUAAUA |
| miR-496 | 457 | 463 | m8 | hsa-miR-496 | AUUACAUGGCCAAUCUC |
| miR-539 | 788 | 794 | m8 | hsa-miR-539 | GGAGAAAUUAUCCUUGGUGUGU |
| miR-544 | 167 | 174 | 1A,m8 | hsa-miR-544 | AUUCUGCAUUUUUAGCAAGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.