Gene Page: HNRNPM
Summary ?
| GeneID | 4670 |
| Symbol | HNRNPM |
| Synonyms | CEAR|HNRNPM4|HNRPM|HNRPM4|HTGR1|NAGR1|hnRNP M |
| Description | heterogeneous nuclear ribonucleoprotein M |
| Reference | MIM:160994|HGNC:HGNC:5046|Ensembl:ENSG00000099783|HPRD:01188|Vega:OTTHUMG00000182383 |
| Gene type | protein-coding |
| Map location | 19p13.2 |
| Pascal p-value | 0.318 |
| Sherlock p-value | 0.245 |
| DEG p-value | DEG:Maycox_2009:CC_BA10_fold_change=1.08:CC_BA10_disease_P=0.0223:HBB_BA9_fold_change=1.15:HBB_BA9_disease_P=0.0368 |
| Fetal beta | 1.175 |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.humanPSD G2Cdb.humanPSP Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DEG:Maycox_2009 | Microarray to determine the expression of over 30000 mRNA transcripts in post-mortem tissue | We included 51 genes whose expression changes are common between two schizophrenia cohorts. |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
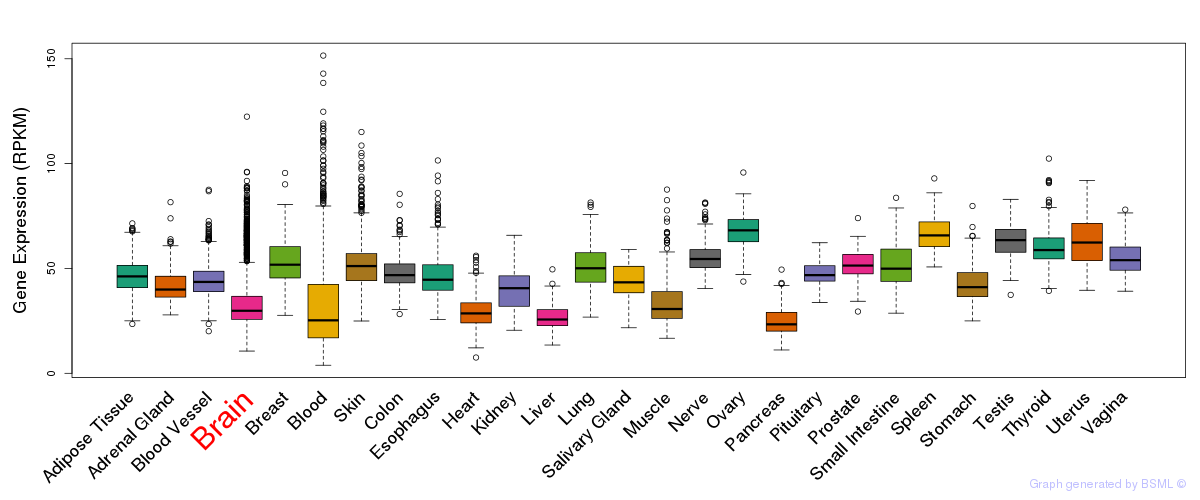
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NDUFS8 | 0.89 | 0.85 |
| C19orf70 | 0.88 | 0.87 |
| AURKAIP1 | 0.88 | 0.84 |
| C16orf13 | 0.87 | 0.84 |
| NDUFA13 | 0.86 | 0.82 |
| C17orf90 | 0.85 | 0.78 |
| TMEM134 | 0.85 | 0.75 |
| FIS1 | 0.85 | 0.75 |
| C7orf59 | 0.85 | 0.78 |
| NDUFA2 | 0.85 | 0.81 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RBM25 | -0.51 | -0.59 |
| UPF2 | -0.50 | -0.49 |
| MYSM1 | -0.49 | -0.50 |
| SFRS12 | -0.49 | -0.56 |
| ZC3H13 | -0.48 | -0.48 |
| BDP1 | -0.47 | -0.46 |
| ATRX | -0.46 | -0.45 |
| ARID4B | -0.46 | -0.46 |
| MAP4K5 | -0.46 | -0.47 |
| ZNF326 | -0.46 | -0.51 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BAT2 | D6S51 | D6S51E | DKFZp686D09175 | G2 | HLA-B associated transcript 2 | - | HPRD,BioGRID | 14667819 |
| CEACAM5 | CD66e | CEA | DKFZp781M2392 | carcinoembryonic antigen-related cell adhesion molecule 5 | - | HPRD | 11406629 |
| GSTK1 | GST13 | glutathione S-transferase kappa 1 | Affinity Capture-MS | BioGRID | 17353931 |
| JUN | AP-1 | AP1 | c-Jun | jun oncogene | - | HPRD,BioGRID | 11823437 |
| LMO3 | DAT1 | MGC26081 | RBTN3 | RBTNL2 | RHOM3 | Rhom-3 | LIM domain only 3 (rhombotin-like 2) | Two-hybrid | BioGRID | 16189514 |
| LYST | CHS | CHS1 | lysosomal trafficking regulator | - | HPRD,BioGRID | 11984006 |
| MAP3K14 | FTDCR1B | HS | HSNIK | NIK | mitogen-activated protein kinase kinase kinase 14 | - | HPRD | 14743216 |
| NFKB1 | DKFZp686C01211 | EBP-1 | KBF1 | MGC54151 | NF-kappa-B | NFKB-p105 | NFKB-p50 | p105 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 | - | HPRD | 14743216 |
| NFKB2 | LYT-10 | LYT10 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) | - | HPRD | 14743216 |
| PHLDA3 | TIH1 | pleckstrin homology-like domain, family A, member 3 | Affinity Capture-MS | BioGRID | 17353931 |
| PTP4A3 | PRL-3 | PRL-R | PRL3 | protein tyrosine phosphatase type IVA, member 3 | Affinity Capture-MS | BioGRID | 17353931 |
| RBM4B | MGC10871 | RBM30 | RBM4L | ZCCHC15 | ZCRB3B | RNA binding motif protein 4B | Two-hybrid | BioGRID | 16189514 |
| RFXANK | ANKRA1 | BLS | F14150_1 | MGC138628 | RFX-B | regulatory factor X-associated ankyrin-containing protein | Affinity Capture-MS | BioGRID | 17353931 |
| TANK | I-TRAF | TRAF2 | TRAF family member-associated NFKB activator | - | HPRD | 14743216 |
| TBK1 | FLJ11330 | NAK | T2K | TANK-binding kinase 1 | - | HPRD | 14743216 |
| TCERG1 | CA150 | MGC133200 | TAF2S | transcription elongation regulator 1 | - | HPRD,BioGRID | 15456888 |
| TSC22D1 | DKFZp686O19206 | MGC17597 | RP11-269C23.2 | TGFB1I4 | TSC22 | TSC22 domain family, member 1 | Affinity Capture-MS | BioGRID | 17353931 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG SPLICEOSOME | 128 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | 140 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA PROCESSING | 161 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA SPLICING | 111 | 58 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS DN | 431 | 263 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER DN | 406 | 230 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LPS UP | 431 | 237 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| OUELLET OVARIAN CANCER INVASIVE VS LMP UP | 117 | 85 | All SZGR 2.0 genes in this pathway |
| OUELLET CULTURED OVARIAN CANCER INVASIVE VS LMP UP | 69 | 40 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 1 DN | 169 | 102 | All SZGR 2.0 genes in this pathway |
| UEDA CENTRAL CLOCK | 88 | 62 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 12 | 36 | 18 | All SZGR 2.0 genes in this pathway |
| TAKAO RESPONSE TO UVB RADIATION DN | 98 | 67 | All SZGR 2.0 genes in this pathway |
| RAMPON ENRICHED LEARNING ENVIRONMENT EARLY UP | 15 | 12 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| MALONEY RESPONSE TO 17AAG DN | 79 | 45 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| ZEMBUTSU SENSITIVITY TO CISPLATIN | 20 | 14 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| STAMBOLSKY TARGETS OF MUTATED TP53 UP | 49 | 35 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |