Gene Page: NOTCH1
Summary ?
| GeneID | 4851 |
| Symbol | NOTCH1 |
| Synonyms | AOS5|AOVD1|TAN1|hN1 |
| Description | notch 1 |
| Reference | MIM:190198|HGNC:HGNC:7881|Ensembl:ENSG00000148400|HPRD:01827|Vega:OTTHUMG00000020935 |
| Gene type | protein-coding |
| Map location | 9q34.3 |
| Pascal p-value | 0.084 |
| Sherlock p-value | 0.719 |
| Fetal beta | 0.871 |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.062 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NOTCH1 | chr9 | 139391864 | G | A | NM_017617 | . | silent | Schizophrenia | DNM:Fromer_2014 |
Section II. Transcriptome annotation
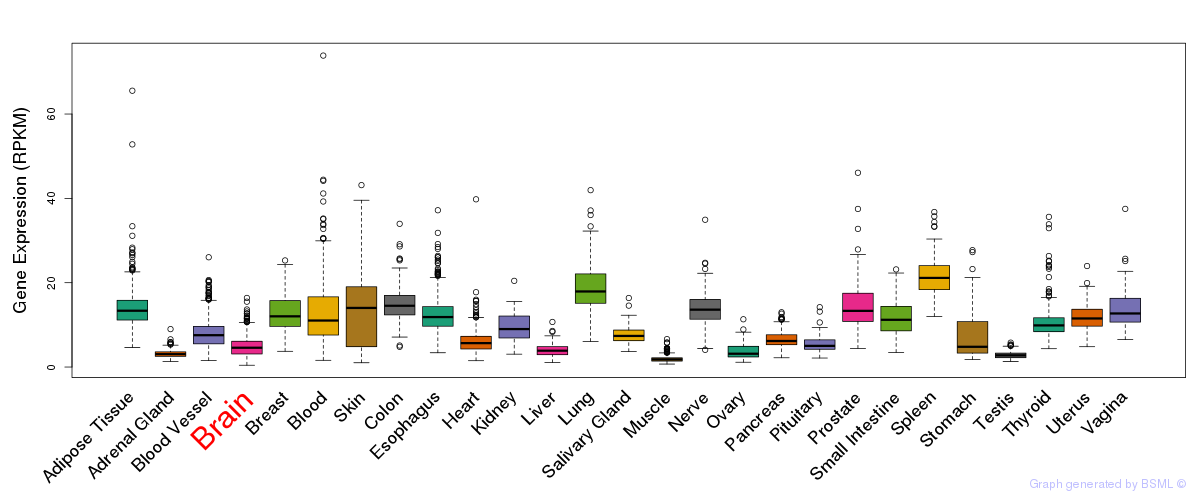
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ZNF384 | 0.95 | 0.96 |
| SAP130 | 0.95 | 0.95 |
| PPIL2 | 0.94 | 0.95 |
| PPRC1 | 0.94 | 0.95 |
| PHF12 | 0.94 | 0.96 |
| POGZ | 0.94 | 0.96 |
| QRICH1 | 0.94 | 0.95 |
| ZNF142 | 0.94 | 0.95 |
| C5orf25 | 0.93 | 0.93 |
| GTF3C2 | 0.93 | 0.95 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.77 | -0.87 |
| MT-CO2 | -0.75 | -0.86 |
| AF347015.27 | -0.75 | -0.85 |
| AF347015.33 | -0.72 | -0.81 |
| IFI27 | -0.72 | -0.82 |
| MT-CYB | -0.72 | -0.82 |
| AF347015.21 | -0.72 | -0.93 |
| C5orf53 | -0.72 | -0.74 |
| AF347015.8 | -0.71 | -0.84 |
| FXYD1 | -0.71 | -0.82 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004872 | receptor activity | IEA | - | |
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 10713164 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0007219 | Notch signaling pathway | TAS | 10713164 | |
| GO:0006955 | immune response | NAS | 1831692 | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0030154 | cell differentiation | IEA | - | |
| GO:0050793 | regulation of developmental process | IEA | - | |
| GO:0045662 | negative regulation of myoblast differentiation | IMP | 10713164 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | TAS | 10713164 | |
| GO:0016021 | integral to membrane | NAS | - | |
| GO:0005886 | plasma membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| APBA1 | D9S411E | MINT1 | X11 | X11A | X11ALPHA | amyloid beta (A4) precursor protein-binding, family A, member 1 | Notch1 interacts with X11-alpha. This interaction was modelled on a demonstrated interaction between mouse Notch1 and human X11-alpha. | BIND | 14756819 |
| CNTN1 | F3 | GP135 | contactin 1 | - | HPRD,BioGRID | 14567914 |
| DLL1 | DELTA1 | DL1 | Delta | delta-like 1 (Drosophila) | Reconstituted Complex | BioGRID | 11006133 |
| DLL4 | MGC126344 | hdelta2 | delta-like 4 (Drosophila) | - | HPRD | 10837024 |11739188 |
| DTX1 | hDx-1 | deltex homolog 1 (Drosophila) | Affinity Capture-Western | BioGRID | 11564735 |
| DTX2 | KIAA1528 | MGC71098 | RNF58 | deltex homolog 2 (Drosophila) | - | HPRD | 11226752 |
| FBXW7 | AGO | CDC4 | DKFZp686F23254 | FBW6 | FBW7 | FBX30 | FBXO30 | FBXW6 | FLJ16457 | SEL-10 | SEL10 | F-box and WD repeat domain containing 7 | - | HPRD,BioGRID | 11425854 |11585921 |
| GSK3B | - | glycogen synthase kinase 3 beta | - | HPRD,BioGRID | 12123574 |
| ITCH | AIF4 | AIP4 | NAPP1 | dJ468O1.1 | itchy E3 ubiquitin protein ligase homolog (mouse) | - | HPRD | 12682059 |
| JAG1 | AGS | AHD | AWS | CD339 | HJ1 | JAGL1 | MGC104644 | jagged 1 (Alagille syndrome) | - | HPRD,BioGRID | 11006133 |
| JAG2 | HJ2 | SER2 | jagged 2 | - | HPRD,BioGRID | 11006133 |
| KAT2B | CAF | P | P/CAF | PCAF | K(lysine) acetyltransferase 2B | - | HPRD | 10747963 |
| LCK | YT16 | p56lck | pp58lck | lymphocyte-specific protein tyrosine kinase | - | HPRD,BioGRID | 14583609 |
| LEF1 | DKFZp586H0919 | TCF1ALPHA | lymphoid enhancer-binding factor 1 | - | HPRD,BioGRID | 11604490 |
| LFNG | SCDO3 | LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase | - | HPRD | 10935626 |12486116 |
| MAML1 | KIAA0200 | Mam-1 | Mam1 | mastermind-like 1 (Drosophila) | - | HPRD,BioGRID | 11101851 |
| MAML2 | DKFZp686N0150 | KIAA1819 | MAM-3 | MAM2 | MAM3 | MGC176701 | MLL-MAML2 | mastermind-like 2 (Drosophila) | Two-hybrid | BioGRID | 12370315 |
| MAML3 | CAGH3 | ERDA3 | GDN | MAM-2 | MAM2 | TNRC3 | mastermind-like 3 (Drosophila) | Two-hybrid | BioGRID | 12370315 |
| MFNG | - | MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase | - | HPRD | 10935626 |12486116 |
| NFKB1 | DKFZp686C01211 | EBP-1 | KBF1 | MGC54151 | NF-kappa-B | NFKB-p105 | NFKB-p50 | p105 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 | - | HPRD,BioGRID | 8642313 |11418662 |
| NOTCH1 | TAN1 | hN1 | Notch homolog 1, translocation-associated (Drosophila) | - | HPRD,BioGRID | 10669757 |
| NOTCH3 | CADASIL | CASIL | Notch homolog 3 (Drosophila) | Phenotypic Suppression | BioGRID | 11404076 |
| NOV | CCN3 | IGFBP9 | nephroblastoma overexpressed gene | - | HPRD,BioGRID | 12050162 |
| NUMB | S171 | numb homolog (Drosophila) | - | HPRD,BioGRID | 8755477 |12682059 |
| PIK3CA | MGC142161 | MGC142163 | PI3K | p110-alpha | phosphoinositide-3-kinase, catalytic, alpha polypeptide | Affinity Capture-Western | BioGRID | 14583609 |
| PIK3CG | PI3CG | PI3K | PI3Kgamma | PIK3 | phosphoinositide-3-kinase, catalytic, gamma polypeptide | - | HPRD | 14583609 |
| PSEN1 | AD3 | FAD | PS1 | S182 | presenilin 1 | Affinity Capture-Western | BioGRID | 10077672 |
| RBPJ | CBF1 | IGKJRB | IGKJRB1 | KBF2 | MGC61669 | RBP-J | RBPJK | RBPSUH | SUH | csl | recombination signal binding protein for immunoglobulin kappa J region | - | HPRD,BioGRID | 8749394 |9111040 |10637481 |
| RELA | MGC131774 | NFKB3 | p65 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | - | HPRD,BioGRID | 8642313 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | - | HPRD,BioGRID | 14638857 |
| SNW1 | Bx42 | MGC119379 | NCOA-62 | PRPF45 | Prp45 | SKIIP | SKIP | SNW domain containing 1 | SKIP interacts with Notch. This interaction was modeled on a demonstrated interaction between human SKIP and mouse Notch. | BIND | 15546612 |
| SNW1 | Bx42 | MGC119379 | NCOA-62 | PRPF45 | Prp45 | SKIIP | SKIP | SNW domain containing 1 | Reconstituted Complex Two-hybrid | BioGRID | 11404076 |
| XRCC6 | CTC75 | CTCBF | G22P1 | KU70 | ML8 | TLAA | X-ray repair complementing defective repair in Chinese hamster cells 6 | Two-hybrid | BioGRID | 16169070 |
| YY1 | DELTA | INO80S | NF-E1 | UCRBP | YIN-YANG-1 | YY1 transcription factor | - | HPRD,BioGRID | 12913000 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG DORSO VENTRAL AXIS FORMATION | 25 | 21 | All SZGR 2.0 genes in this pathway |
| KEGG NOTCH SIGNALING PATHWAY | 47 | 35 | All SZGR 2.0 genes in this pathway |
| KEGG PRION DISEASES | 35 | 28 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PS1 PATHWAY | 14 | 14 | All SZGR 2.0 genes in this pathway |
| PID NOTCH PATHWAY | 59 | 49 | All SZGR 2.0 genes in this pathway |
| PID PS1 PATHWAY | 46 | 39 | All SZGR 2.0 genes in this pathway |
| PID DELTA NP63 PATHWAY | 47 | 34 | All SZGR 2.0 genes in this pathway |
| PID HES HEY PATHWAY | 48 | 39 | All SZGR 2.0 genes in this pathway |
| PYEON HPV POSITIVE TUMORS UP | 98 | 47 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS UP | 332 | 228 | All SZGR 2.0 genes in this pathway |
| LEE NEURAL CREST STEM CELL DN | 118 | 79 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION KRAS DN | 142 | 95 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 DN | 229 | 142 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION 12HR | 43 | 35 | All SZGR 2.0 genes in this pathway |
| LUND SILENCED BY METHYLATION | 16 | 12 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 UP | 209 | 139 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 AND HIF1A DN | 103 | 71 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| MORI MATURE B LYMPHOCYTE DN | 75 | 43 | All SZGR 2.0 genes in this pathway |
| MA MYELOID DIFFERENTIATION DN | 44 | 30 | All SZGR 2.0 genes in this pathway |
| PARK HSC MARKERS | 44 | 31 | All SZGR 2.0 genes in this pathway |
| LEE AGING CEREBELLUM UP | 84 | 58 | All SZGR 2.0 genes in this pathway |
| MOREIRA RESPONSE TO TSA DN | 18 | 16 | All SZGR 2.0 genes in this pathway |
| SAFFORD T LYMPHOCYTE ANERGY | 87 | 54 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN | 409 | 268 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| CAMPS COLON CANCER COPY NUMBER UP | 92 | 45 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| RAY TUMORIGENESIS BY ERBB2 CDC25A DN | 159 | 105 | All SZGR 2.0 genes in this pathway |
| LU TUMOR ANGIOGENESIS UP | 25 | 22 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS DN | 138 | 92 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| BRUNEAU HEART GREAT VESSELS AND VALVULOGENESIS | 8 | 8 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| FONTAINE FOLLICULAR THYROID ADENOMA DN | 68 | 45 | All SZGR 2.0 genes in this pathway |
| FONTAINE PAPILLARY THYROID CARCINOMA UP | 66 | 38 | All SZGR 2.0 genes in this pathway |
| FUKUSHIMA TNFSF11 TARGETS | 16 | 14 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS DN | 213 | 127 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| WANG NFKB TARGETS | 25 | 15 | All SZGR 2.0 genes in this pathway |
| DUAN PRDM5 TARGETS | 79 | 52 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| WANG THOC1 TARGETS DN | 20 | 14 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE VIA P38 COMPLETE | 227 | 151 | All SZGR 2.0 genes in this pathway |
| SMIRNOV RESPONSE TO IR 6HR UP | 166 | 97 | All SZGR 2.0 genes in this pathway |
| WARTERS RESPONSE TO IR SKIN | 83 | 44 | All SZGR 2.0 genes in this pathway |
| WARTERS IR RESPONSE 5GY | 47 | 23 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-139 | 1559 | 1566 | 1A,m8 | hsa-miR-139brain | UCUACAGUGCACGUGUCU |
| miR-25/32/92/363/367 | 1602 | 1608 | 1A | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| miR-30-5p | 1520 | 1526 | 1A | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-34/449 | 180 | 186 | m8 | hsa-miR-34abrain | UGGCAGUGUCUUAGCUGGUUGUU |
| hsa-miR-34c | AGGCAGUGUAGUUAGCUGAUUGC | ||||
| hsa-miR-449 | UGGCAGUGUAUUGUUAGCUGGU | ||||
| hsa-miR-449b | AGGCAGUGUAUUGUUAGCUGGC | ||||
| hsa-miR-34abrain | UGGCAGUGUCUUAGCUGGUUGUU | ||||
| hsa-miR-34c | AGGCAGUGUAGUUAGCUGAUUGC | ||||
| hsa-miR-449 | UGGCAGUGUAUUGUUAGCUGGU | ||||
| hsa-miR-449b | AGGCAGUGUAUUGUUAGCUGGC | ||||
| miR-34b | 181 | 187 | m8 | hsa-miR-34b | UAGGCAGUGUCAUUAGCUGAUUG |
| hsa-miR-34b | UAGGCAGUGUCAUUAGCUGAUUG | ||||
| miR-495 | 1588 | 1594 | 1A | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.