Gene Page: BARD1
Summary ?
| GeneID | 580 |
| Symbol | BARD1 |
| Synonyms | - |
| Description | BRCA1 associated RING domain 1 |
| Reference | MIM:601593|HGNC:HGNC:952|Ensembl:ENSG00000138376|HPRD:03354|Vega:OTTHUMG00000133016 |
| Gene type | protein-coding |
| Map location | 2q35 |
| Pascal p-value | 0.26 |
| Sherlock p-value | 0.701 |
| Fetal beta | 1.216 |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00916 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01016 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.3518 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs9789570 | chr2 | 215694649 | BARD1 | 580 | 0.01 | cis | ||
| rs2166458 | chr2 | 215716168 | BARD1 | 580 | 0 | cis | ||
| rs777307 | chr2 | 215731208 | BARD1 | 580 | 0.18 | cis | ||
| rs7578759 | chr2 | 215798407 | BARD1 | 580 | 0.17 | cis | ||
| rs1409612 | chr2 | 215800709 | BARD1 | 580 | 0.03 | cis | ||
| rs2166458 | chr2 | 215716168 | BARD1 | 580 | 0.11 | trans | ||
| rs1007573 | chr12 | 99092620 | BARD1 | 580 | 0.13 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
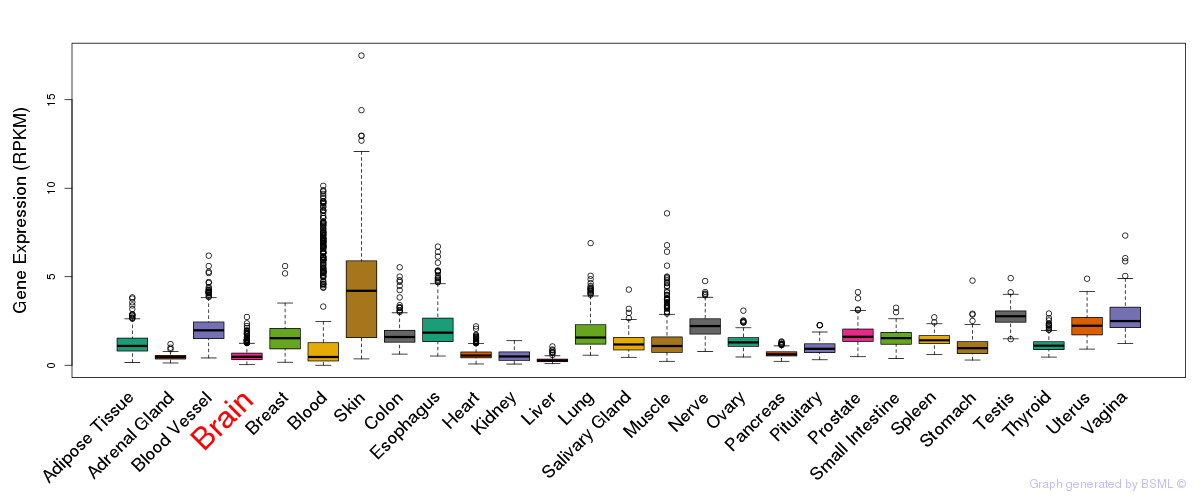
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| USP40 | 0.79 | 0.82 |
| CAPN2 | 0.74 | 0.70 |
| SLC44A2 | 0.74 | 0.70 |
| AK3 | 0.72 | 0.69 |
| MCCC2 | 0.72 | 0.69 |
| CHD1L | 0.72 | 0.72 |
| BMP7 | 0.72 | 0.70 |
| ALDH7A1 | 0.72 | 0.72 |
| CPT2 | 0.71 | 0.72 |
| SEPT2 | 0.71 | 0.74 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| IL32 | -0.40 | -0.37 |
| FAM159B | -0.38 | -0.26 |
| AF347015.21 | -0.36 | -0.18 |
| RP9P | -0.32 | -0.24 |
| TTC9B | -0.31 | -0.28 |
| CLEC3B | -0.31 | -0.36 |
| AL050337.1 | -0.31 | -0.13 |
| AC087071.1 | -0.30 | -0.13 |
| AL049792.1 | -0.29 | -0.15 |
| AC021914.1 | -0.29 | -0.10 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003723 | RNA binding | IDA | 12419249 | |
| GO:0005515 | protein binding | IPI | 8944023 |10026184 |10477523 |15184379 |15265711 |15782130 | |
| GO:0004842 | ubiquitin-protein ligase activity | NAS | 15905410 | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0019900 | kinase binding | NAS | 15782130 | |
| GO:0042803 | protein homodimerization activity | IPI | 15265711 | |
| GO:0046872 | metal ion binding | IEA | - | |
| GO:0046982 | protein heterodimerization activity | IPI | 15265711 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001894 | tissue homeostasis | TAS | 15782130 | |
| GO:0007050 | cell cycle arrest | NAS | 15632137 | |
| GO:0006974 | response to DNA damage stimulus | NAS | 15905410 | |
| GO:0043065 | positive regulation of apoptosis | IMP | 15782130 | |
| GO:0016567 | protein ubiquitination | NAS | 15905410 | |
| GO:0043066 | negative regulation of apoptosis | IMP | 15265711 | |
| GO:0042325 | regulation of phosphorylation | IMP | 15782130 | |
| GO:0031441 | negative regulation of mRNA 3'-end processing | NAS | 15905410 | |
| GO:0045732 | positive regulation of protein catabolic process | NAS | 15905410 | |
| GO:0046826 | negative regulation of protein export from nucleus | IDA | 15265711 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000151 | ubiquitin ligase complex | NAS | 14976165 | |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005634 | nucleus | IDA | 15265711 | |
| GO:0005634 | nucleus | IMP | 15632137 | |
| GO:0005737 | cytoplasm | IDA | 15265711 | |
| GO:0031436 | BRCA1-BARD1 complex | IDA | 15265711 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BARD1 | - | BRCA1 associated RING domain 1 | Reconstituted Complex | BioGRID | 11257228 |
| BCL3 | BCL4 | D19S37 | B-cell CLL/lymphoma 3 | - | HPRD,BioGRID | 10362352 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | Affinity Capture-MS Affinity Capture-Western Co-crystal Structure Co-localization Co-purification FRET Reconstituted Complex Two-hybrid | BioGRID | 8944023 |9342365 |9738006 |9788437 |10026184 |10477523 |10635334 |11257228 |11278247 |11301010 |11498787 |11504724 |11526114 |11573085 |11773071 |11925436 |11927591 |12431996 |12438698 |12485996 |12700228 |12732733 |12887909 |12890688 |12951069 |14636569 |14638690 |14647430 |15159397 |15184379 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | - | HPRD | 8944023 |10026184 |11573085 |11773071 |12431996 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | - | HPRD | 8944023 |10026184 |11773071 |12431996 |
| BRCA2 | BRCC2 | FACD | FAD | FAD1 | FANCB | FANCD | FANCD1 | breast cancer 2, early onset | Co-purification Reconstituted Complex | BioGRID | 14636569 |
| BRCC3 | BRCC36 | C6.1A | CXorf53 | BRCA1/BRCA2-containing complex, subunit 3 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 14636569 |
| BRE | BRCC4 | BRCC45 | brain and reproductive organ-expressed (TNFRSF1A modulator) | Affinity Capture-MS Affinity Capture-Western | BioGRID | 14636569 |
| CSTF1 | CstF-50 | CstFp50 | cleavage stimulation factor, 3' pre-RNA, subunit 1, 50kDa | - | HPRD,BioGRID | 10477523 |11257228 |
| CSTF2 | CstF-64 | cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa | Affinity Capture-Western Co-purification | BioGRID | 10477523 |11257228 |
| E2F1 | E2F-1 | RBAP1 | RBBP3 | RBP3 | E2F transcription factor 1 | E2F1 interacts with the BARD1 promoter. | BIND | 11799067 |
| E2F4 | E2F-4 | E2F transcription factor 4, p107/p130-binding | E2F4 interacts with the BARD1 promoter region. | BIND | 11799067 |
| EWSR1 | EWS | Ewing sarcoma breakpoint region 1 | - | HPRD,BioGRID | 12183411 |
| FANCD2 | DKFZp762A223 | FA-D2 | FA4 | FACD | FAD | FAD2 | FANCD | FLJ23826 | Fanconi anemia, complementation group D2 | Co-localization Reconstituted Complex | BioGRID | 12887909 |
| H2AFX | H2A.X | H2A/X | H2AX | H2A histone family, member X | Reconstituted Complex | BioGRID | 11927591 |12485996 |
| HIST2H2AB | - | histone cluster 2, H2ab | Reconstituted Complex | BioGRID | 11927591 |
| ING5 | FLJ23842 | p28ING5 | inhibitor of growth family, member 5 | - | HPRD | 15383276 |
| MED17 | CRSP6 | CRSP77 | DRIP80 | FLJ10812 | TRAP80 | mediator complex subunit 17 | Co-purification | BioGRID | 12154023 |
| MSH2 | COCA1 | FCC1 | HNPCC | HNPCC1 | LCFS2 | mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli) | - | HPRD,BioGRID | 11498787 |
| MSH3 | DUP | MGC163306 | MGC163308 | MRP1 | mutS homolog 3 (E. coli) | Reconstituted Complex | BioGRID | 11498787 |
| MSH6 | GTBP | HNPCC5 | HSAP | mutS homolog 6 (E. coli) | Reconstituted Complex | BioGRID | 11498787 |
| NFKBIA | IKBA | MAD-3 | NFKBI | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | Two-hybrid | BioGRID | 10362352 |
| NPM1 | B23 | MGC104254 | NPM | nucleophosmin (nucleolar phosphoprotein B23, numatrin) | Affinity Capture-MS Affinity Capture-Western Reconstituted Complex | BioGRID | 15184379 |
| POLR2A | MGC75453 | POLR2 | POLRA | RPB1 | RPBh1 | RPO2 | RPOL2 | RpIILS | hRPB220 | hsRPB1 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | - | HPRD | 12154023 |
| POU2F1 | OCT1 | OTF1 | POU class 2 homeobox 1 | - | HPRD | 11777930 |
| RAD51 | BRCC5 | HRAD51 | HsRad51 | HsT16930 | RAD51A | RECA | RAD51 homolog (RecA homolog, E. coli) (S. cerevisiae) | Affinity Capture-MS Affinity Capture-Western | BioGRID | 14636569 |
| RBBP8 | CTIP | RIM | retinoblastoma binding protein 8 | - | HPRD,BioGRID | 10764811 |
| RELA | MGC131774 | NFKB3 | p65 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | Affinity Capture-Western | BioGRID | 12700228 |
| TCERG1 | CA150 | MGC133200 | TAF2S | transcription elongation regulator 1 | - | HPRD | 15383276 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 14636569 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | TP53 (p53) interacts with BARD1. | BIND | 15782130 |
| UBA1 | A1S9 | A1S9T | A1ST | AMCX1 | GXP1 | MGC4781 | SMAX2 | UBA1A | UBE1 | UBE1X | ubiquitin-like modifier activating enzyme 1 | Reconstituted Complex | BioGRID | 11278247 |11927591 |12431996 |12438698 |12485996 |12887909 |15184379 |
| UBE2D1 | E2(17)KB1 | SFT | UBC4/5 | UBCH5 | UBCH5A | ubiquitin-conjugating enzyme E2D 1 (UBC4/5 homolog, yeast) | Reconstituted Complex | BioGRID | 11278247 |11927591 |12438698 |12485996 |12887909 |12890688 |14636569 |15184379 |
| UBE2D2 | E2(17)KB2 | PUBC1 | UBC4 | UBC4/5 | UBCH5B | ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast) | Reconstituted Complex | BioGRID | 12431996 |
| UBE2D3 | E2(17)KB3 | MGC43926 | MGC5416 | UBC4/5 | UBCH5C | ubiquitin-conjugating enzyme E2D 3 (UBC4/5 homolog, yeast) | - | HPRD | 12732733 |
| UBE2L3 | E2-F1 | L-UBC | UBCH7 | UbcM4 | ubiquitin-conjugating enzyme E2L 3 | - | HPRD | 12732733 |
| UBE3A | ANCR | AS | E6-AP | EPVE6AP | FLJ26981 | HPVE6A | ubiquitin protein ligase E3A | Reconstituted Complex | BioGRID | 12890688 |
| XIST | DKFZp779P0129 | DXS1089 | DXS399E | NCRNA00001 | SXI1 | XCE | XIC | swd66 | X (inactive)-specific transcript (non-protein coding) | Co-localization | BioGRID | 12419249 |
| XRCC6 | CTC75 | CTCBF | G22P1 | KU70 | ML8 | TLAA | X-ray repair complementing defective repair in Chinese hamster cells 6 | BARD1 interacts with XRCC6 (Ku70). | BIND | 15782130 |
| ZHX1 | - | zinc fingers and homeoboxes 1 | - | HPRD | 15383276 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID BARD1 PATHWAY | 29 | 19 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR MARKERS UP | 21 | 13 | All SZGR 2.0 genes in this pathway |
| PYEON HPV POSITIVE TUMORS UP | 98 | 47 | All SZGR 2.0 genes in this pathway |
| CHEMNITZ RESPONSE TO PROSTAGLANDIN E2 UP | 146 | 86 | All SZGR 2.0 genes in this pathway |
| CORRE MULTIPLE MYELOMA DN | 62 | 41 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY UP | 430 | 232 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS TURQUOISE DN | 53 | 25 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY DN | 362 | 238 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN | 536 | 332 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN | 459 | 276 | All SZGR 2.0 genes in this pathway |
| HAHTOLA SEZARY SYNDROM UP | 98 | 58 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES SKIN UP | 177 | 113 | All SZGR 2.0 genes in this pathway |
| TANG SENESCENCE TP53 TARGETS DN | 57 | 33 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION HBZ | 41 | 27 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| KIM MYCN AMPLIFICATION TARGETS DN | 103 | 59 | All SZGR 2.0 genes in this pathway |
| PUJANA BREAST CANCER LIT INT NETWORK | 101 | 73 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS | 539 | 324 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| NELSON RESPONSE TO ANDROGEN DN | 19 | 14 | All SZGR 2.0 genes in this pathway |
| PENG GLUTAMINE DEPRIVATION UP | 38 | 25 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA DN | 289 | 166 | All SZGR 2.0 genes in this pathway |
| POMEROY MEDULLOBLASTOMA PROGNOSIS DN | 43 | 28 | All SZGR 2.0 genes in this pathway |
| REN BOUND BY E2F | 61 | 40 | All SZGR 2.0 genes in this pathway |
| SASAKI ADULT T CELL LEUKEMIA | 176 | 122 | All SZGR 2.0 genes in this pathway |
| MOREAUX B LYMPHOCYTE MATURATION BY TACI DN | 73 | 45 | All SZGR 2.0 genes in this pathway |
| FERRANDO T ALL WITH MLL ENL FUSION DN | 87 | 57 | All SZGR 2.0 genes in this pathway |
| HADDAD B LYMPHOCYTE PROGENITOR | 293 | 193 | All SZGR 2.0 genes in this pathway |
| VERNELL RETINOBLASTOMA PATHWAY UP | 70 | 47 | All SZGR 2.0 genes in this pathway |
| ZHAN V2 LATE DIFFERENTIATION GENES | 45 | 34 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI DN | 172 | 107 | All SZGR 2.0 genes in this pathway |
| LY AGING PREMATURE DN | 30 | 17 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 2 | 50 | 34 | All SZGR 2.0 genes in this pathway |
| KANG DOXORUBICIN RESISTANCE UP | 54 | 33 | All SZGR 2.0 genes in this pathway |
| SONG TARGETS OF IE86 CMV PROTEIN | 60 | 42 | All SZGR 2.0 genes in this pathway |
| LY AGING OLD DN | 56 | 35 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1 | 419 | 273 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| SARRIO EPITHELIAL MESENCHYMAL TRANSITION UP | 180 | 114 | All SZGR 2.0 genes in this pathway |
| DE YY1 TARGETS DN | 92 | 64 | All SZGR 2.0 genes in this pathway |
| DAIRKEE CANCER PRONE RESPONSE BPA E2 | 118 | 65 | All SZGR 2.0 genes in this pathway |
| TOYOTA TARGETS OF MIR34B AND MIR34C | 463 | 262 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR UP | 294 | 199 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| FRASOR RESPONSE TO SERM OR FULVESTRANT DN | 50 | 29 | All SZGR 2.0 genes in this pathway |
| RUIZ TNC TARGETS DN | 142 | 79 | All SZGR 2.0 genes in this pathway |
| CHANG CYCLING GENES | 148 | 83 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G123 UP | 45 | 21 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS PROLIFERATION UP | 178 | 108 | All SZGR 2.0 genes in this pathway |
| KOBAYASHI EGFR SIGNALING 24HR DN | 251 | 151 | All SZGR 2.0 genes in this pathway |
| FOURNIER ACINAR DEVELOPMENT LATE 2 | 277 | 172 | All SZGR 2.0 genes in this pathway |
| PYEON CANCER HEAD AND NECK VS CERVICAL UP | 193 | 95 | All SZGR 2.0 genes in this pathway |
| HAHTOLA CTCL CUTANEOUS | 26 | 19 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| ZEMBUTSU SENSITIVITY TO VINCRISTINE | 19 | 10 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 4HR DN | 254 | 158 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G1 S | 147 | 76 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR UP | 324 | 193 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |