Gene Page: BLM
Summary ?
| GeneID | 641 |
| Symbol | BLM |
| Synonyms | BS|RECQ2|RECQL2|RECQL3 |
| Description | Bloom syndrome RecQ like helicase |
| Reference | MIM:604610|HGNC:HGNC:1058|Ensembl:ENSG00000197299|HPRD:05211|Vega:OTTHUMG00000149834 |
| Gene type | protein-coding |
| Map location | 15q26.1 |
| Pascal p-value | 0.011 |
| Fetal beta | 0.094 |
| DMG | 1 (# studies) |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg07973395 | 15 | 91260024 | BLM | 6.16E-9 | -0.014 | 3.26E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
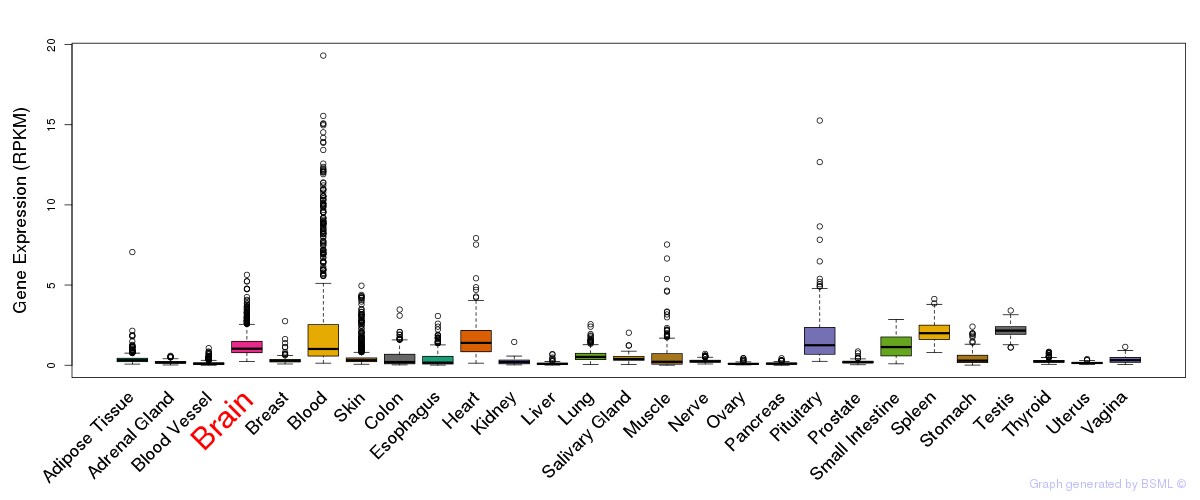
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PSMD13 | 0.87 | 0.86 |
| EIF2B3 | 0.85 | 0.85 |
| CAMLG | 0.85 | 0.87 |
| PSMB7 | 0.85 | 0.87 |
| OCIAD2 | 0.84 | 0.84 |
| CCT7 | 0.84 | 0.84 |
| COQ3 | 0.84 | 0.86 |
| EIF5A | 0.84 | 0.84 |
| EAPP | 0.84 | 0.85 |
| ZNF622 | 0.83 | 0.83 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CYB | -0.72 | -0.75 |
| AF347015.33 | -0.71 | -0.73 |
| AF347015.26 | -0.71 | -0.76 |
| MT-CO2 | -0.71 | -0.70 |
| AF347015.8 | -0.71 | -0.73 |
| AF347015.15 | -0.71 | -0.75 |
| AF347015.2 | -0.69 | -0.73 |
| AF347015.27 | -0.69 | -0.71 |
| AF347015.31 | -0.68 | -0.70 |
| AF347015.9 | -0.67 | -0.73 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ATM | AT1 | ATA | ATC | ATD | ATDC | ATE | DKFZp781A0353 | MGC74674 | TEL1 | TELO1 | ataxia telangiectasia mutated | Affinity Capture-Western Co-purification Reconstituted Complex | BioGRID | 10783165 |12034743 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | Co-purification | BioGRID | 10783165 |
| CASP3 | CPP32 | CPP32B | SCA-1 | caspase 3, apoptosis-related cysteine peptidase | Biochemical Activity | BioGRID | 11470874 |
| CHAF1A | CAF-1 | CAF1 | CAF1B | CAF1P150 | MGC71229 | P150 | chromatin assembly factor 1, subunit A (p150) | Affinity Capture-Western Far Western Two-hybrid | BioGRID | 15143166 |
| CHEK1 | CHK1 | CHK1 checkpoint homolog (S. pombe) | Affinity Capture-Western Co-localization | BioGRID | 15364958 |
| FANCA | FA | FA-H | FA1 | FAA | FACA | FAH | FANCH | MGC75158 | Fanconi anemia, complementation group A | Affinity Capture-MS | BioGRID | 12973351 |
| FANCD2 | DKFZp762A223 | FA-D2 | FA4 | FACD | FAD | FAD2 | FANCD | FLJ23826 | Fanconi anemia, complementation group D2 | Affinity Capture-Western | BioGRID | 15257300 |
| FEN1 | FEN-1 | MF1 | RAD2 | flap structure-specific endonuclease 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 14688284 |
| H2AFX | H2A.X | H2A/X | H2AX | H2A histone family, member X | Affinity Capture-Western Co-localization | BioGRID | 15364958 |
| MLH1 | COCA2 | FCC2 | HNPCC | HNPCC2 | MGC5172 | hMLH1 | mutL homolog 1, colon cancer, nonpolyposis type 2 (E. coli) | - | HPRD,BioGRID | 11691925 |
| MLH1 | COCA2 | FCC2 | HNPCC | HNPCC2 | MGC5172 | hMLH1 | mutL homolog 1, colon cancer, nonpolyposis type 2 (E. coli) | MLH1 interacts with BLM. | BIND | 11691925 |
| MRE11A | ATLD | HNGS1 | MRE11 | MRE11B | MRE11 meiotic recombination 11 homolog A (S. cerevisiae) | Co-purification | BioGRID | 10783165 |
| MSH2 | COCA1 | FCC1 | HNPCC | HNPCC1 | LCFS2 | mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli) | Co-purification | BioGRID | 10783165 |
| MSH6 | GTBP | HNPCC5 | HSAP | mutS homolog 6 (E. coli) | Co-purification | BioGRID | 10783165 |
| MX1 | IFI-78K | IFI78 | MX | MxA | myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) | - | HPRD,BioGRID | 11716541 |
| NBN | AT-V1 | AT-V2 | ATV | FLJ10155 | MGC87362 | NBS | NBS1 | P95 | nibrin | Co-purification | BioGRID | 10783165 |
| RAD50 | RAD50-2 | hRad50 | RAD50 homolog (S. cerevisiae) | Co-purification | BioGRID | 10783165 |
| RAD51 | BRCC5 | HRAD51 | HsRad51 | HsT16930 | RAD51A | RECA | RAD51 homolog (RecA homolog, E. coli) (S. cerevisiae) | - | HPRD,BioGRID | 11278509 |
| RAD51 | BRCC5 | HRAD51 | HsRad51 | HsT16930 | RAD51A | RECA | RAD51 homolog (RecA homolog, E. coli) (S. cerevisiae) | BLM interacts with RAD51. | BIND | 11278509 |
| RAD51L3 | HsTRAD | R51H3 | RAD51D | Trad | RAD51-like 3 (S. cerevisiae) | - | HPRD,BioGRID | 12975363 |
| RFC1 | A1 | MGC51786 | MHCBFB | PO-GA | RECC1 | RFC | RFC140 | replication factor C (activator 1) 1, 145kDa | Co-purification | BioGRID | 10783165 |
| RPA1 | HSSB | REPA1 | RF-A | RP-A | RPA70 | replication protein A1, 70kDa | Biochemical Activity Co-localization Reconstituted Complex | BioGRID | 10825162 |11950880 |12181313 |
| SMC1A | CDLS2 | DKFZp686L19178 | DXS423E | KIAA0178 | MGC138332 | SB1.8 | SMC1 | SMC1L1 | SMC1alpha | SMCB | structural maintenance of chromosomes 1A | - | HPRD | 11877377 |
| TERF2 | TRBF2 | TRF2 | telomeric repeat binding factor 2 | - | HPRD,BioGRID | 12181313 |
| TERF2 | TRBF2 | TRF2 | telomeric repeat binding factor 2 | BLM interacts with TRF2. | BIND | 12181313 |
| TOP3A | TOP3 | topoisomerase (DNA) III alpha | BLM interacts with Top3A (Topo III-alpha). | BIND | 15775963 |
| TOP3A | TOP3 | topoisomerase (DNA) III alpha | - | HPRD | 10734115 |11406610 |
| TOP3A | TOP3 | topoisomerase (DNA) III alpha | BLM interacts with Topoisomerase III-alpha. | BIND | 10734115 |
| TOP3A | TOP3 | topoisomerase (DNA) III alpha | Affinity Capture-Western Co-localization Far Western Reconstituted Complex | BioGRID | 10734115 |10825162 |11406610 |11470874 |15143166 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | BLM interacts with p53. | BIND | 12080066 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | BLM interacts with p53. This interaction was modeled on a demonstrated interaction between BLM and p53 both from African green monkey. | BIND | 15806145 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | - | HPRD,BioGRID | 11781842 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | p53 interacts with BLM. | BIND | 11781842 |
| TP53BP1 | 53BP1 | FLJ41424 | MGC138366 | p202 | tumor protein p53 binding protein 1 | Affinity Capture-Western Co-localization | BioGRID | 15364958 |
| WRN | DKFZp686C2056 | RECQ3 | RECQL2 | RECQL3 | Werner syndrome | BLM interacts with WRN. | BIND | 11919194 |
| WRN | DKFZp686C2056 | RECQ3 | RECQL2 | RECQL3 | Werner syndrome | - | HPRD,BioGRID | 11919194 |
| XRCC2 | DKFZp781P0919 | X-ray repair complementing defective repair in Chinese hamster cells 2 | Affinity Capture-Western Reconstituted Complex | BioGRID | 12975363 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG HOMOLOGOUS RECOMBINATION | 28 | 17 | All SZGR 2.0 genes in this pathway |
| PID FANCONI PATHWAY | 47 | 28 | All SZGR 2.0 genes in this pathway |
| PID ATM PATHWAY | 34 | 25 | All SZGR 2.0 genes in this pathway |
| PID TELOMERASE PATHWAY | 68 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME MEIOSIS | 116 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME MEIOTIC RECOMBINATION | 86 | 62 | All SZGR 2.0 genes in this pathway |
| SOTIRIOU BREAST CANCER GRADE 1 VS 3 UP | 151 | 84 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION | 77 | 51 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| FURUKAWA DUSP6 TARGETS PCI35 DN | 74 | 40 | All SZGR 2.0 genes in this pathway |
| PUJANA BREAST CANCER LIT INT NETWORK | 101 | 73 | All SZGR 2.0 genes in this pathway |
| PUJANA XPRSS INT NETWORK | 168 | 103 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA CENTERED NETWORK | 117 | 72 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN MELANOMA RELAPSE UP | 61 | 25 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| FERREIRA EWINGS SARCOMA UNSTABLE VS STABLE UP | 167 | 92 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPLICATION GENES | 147 | 87 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL AND BRAIN QTL TRANS | 185 | 114 | All SZGR 2.0 genes in this pathway |
| PEART HDAC PROLIFERATION CLUSTER UP | 57 | 35 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 1 DN | 163 | 115 | All SZGR 2.0 genes in this pathway |
| VERNELL RETINOBLASTOMA PATHWAY UP | 70 | 47 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE DN | 123 | 76 | All SZGR 2.0 genes in this pathway |
| LINDVALL IMMORTALIZED BY TERT DN | 80 | 56 | All SZGR 2.0 genes in this pathway |
| SONG TARGETS OF IE86 CMV PROTEIN | 60 | 42 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| SARRIO EPITHELIAL MESENCHYMAL TRANSITION UP | 180 | 114 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN DN | 249 | 165 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| BOHN PRIMARY IMMUNODEFICIENCY SYNDROM UP | 47 | 30 | All SZGR 2.0 genes in this pathway |
| FRASOR RESPONSE TO SERM OR FULVESTRANT DN | 50 | 29 | All SZGR 2.0 genes in this pathway |
| RUIZ TNC TARGETS DN | 142 | 79 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER ESR1 DN | 240 | 153 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR DN | 185 | 116 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR DN | 277 | 166 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS DN | 89 | 50 | All SZGR 2.0 genes in this pathway |
| POOLA INVASIVE BREAST CANCER UP | 288 | 168 | All SZGR 2.0 genes in this pathway |
| PUJANA BREAST CANCER WITH BRCA1 MUTATED UP | 56 | 27 | All SZGR 2.0 genes in this pathway |
| KOBAYASHI EGFR SIGNALING 24HR DN | 251 | 151 | All SZGR 2.0 genes in this pathway |
| FOURNIER ACINAR DEVELOPMENT LATE 2 | 277 | 172 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| WONG EMBRYONIC STEM CELL CORE | 335 | 193 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE S | 162 | 86 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA CLASSICAL | 162 | 122 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 DN | 374 | 217 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR UP | 324 | 193 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
| ALFANO MYC TARGETS | 239 | 156 | All SZGR 2.0 genes in this pathway |