Gene Page: SMARCA4
Summary ?
| GeneID | 6597 |
| Symbol | SMARCA4 |
| Synonyms | BAF190|BAF190A|BRG1|MRD16|RTPS2|SNF2|SNF2L4|SNF2LB|SWI2|hSNF2b |
| Description | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| Reference | MIM:603254|HGNC:HGNC:11100|Ensembl:ENSG00000127616|HPRD:04459|Vega:OTTHUMG00000169272 |
| Gene type | protein-coding |
| Map location | 19p13.2 |
| Pascal p-value | 0.598 |
| Sherlock p-value | 0.021 |
| Fetal beta | 0.751 |
| DMG | 2 (# studies) |
| Support | CompositeSet Darnell FMRP targets Ascano FMRP targets Chromatin Remodeling Genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 2 |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 2 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 3 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0172 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg12411068 | 19 | 11072235 | SMARCA4 | -0.048 | 0.27 | DMG:Nishioka_2013 | |
| cg25397562 | 19 | 11616711 | SMARCA4 | 0.001 | 3.654 | DMG:vanEijk_2014 |
Section II. Transcriptome annotation
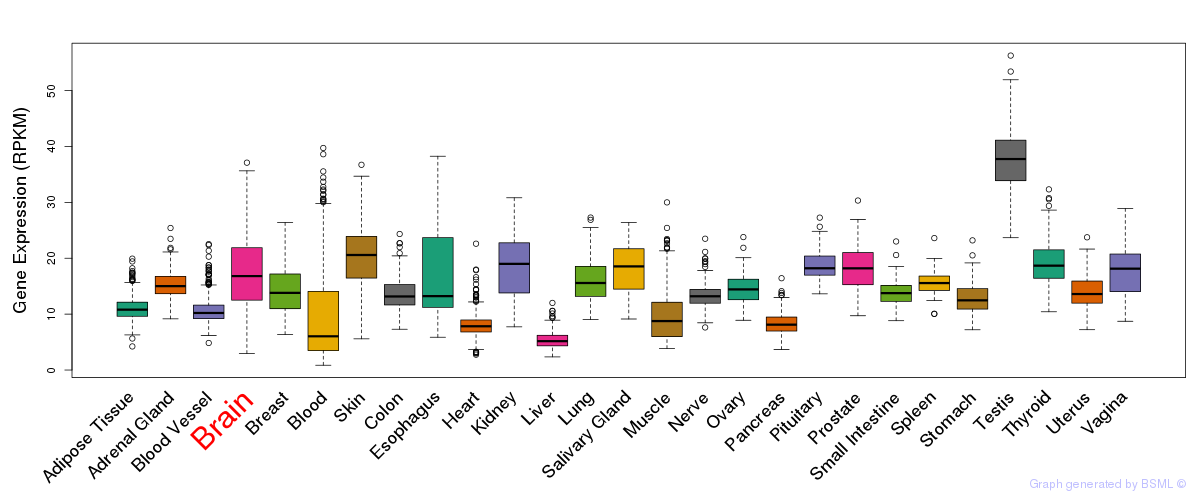
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TPRKB | 0.89 | 0.81 |
| CETN3 | 0.85 | 0.76 |
| RPL35A | 0.84 | 0.83 |
| CCDC58 | 0.81 | 0.68 |
| PSMA2 | 0.81 | 0.73 |
| SNRPE | 0.81 | 0.66 |
| TBCA | 0.81 | 0.73 |
| RPL23 | 0.80 | 0.82 |
| RPL24 | 0.80 | 0.76 |
| TPT1 | 0.79 | 0.77 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ZBTB47 | -0.54 | -0.63 |
| TMEM184B | -0.53 | -0.68 |
| PK4P | -0.53 | -0.64 |
| BCL2L2 | -0.52 | -0.69 |
| ATP10A | -0.52 | -0.65 |
| VASN | -0.51 | -0.65 |
| GAS2L1 | -0.50 | -0.57 |
| TNS1 | -0.50 | -0.70 |
| SYNM | -0.50 | -0.67 |
| PEAR1 | -0.50 | -0.64 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003677 | DNA binding | IEA | - | |
| GO:0003682 | chromatin binding | IEA | - | |
| GO:0003700 | transcription factor activity | TAS | 8208605 | |
| GO:0003713 | transcription coactivator activity | TAS | 8208605 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004386 | helicase activity | IEA | - | |
| GO:0004386 | helicase activity | TAS | 8208605 | |
| GO:0016787 | hydrolase activity | IEA | - | |
| GO:0008134 | transcription factor binding | IEA | - | |
| GO:0042802 | identical protein binding | IPI | 12917342 | |
| GO:0047485 | protein N-terminus binding | IPI | 12917342 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007403 | glial cell fate determination | IEA | Glial (GO term level: 10) | - |
| GO:0030902 | hindbrain development | IEA | Brain (GO term level: 8) | - |
| GO:0030900 | forebrain development | IEA | Brain (GO term level: 8) | - |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | IEA | - | |
| GO:0001835 | blastocyst hatching | IEA | - | |
| GO:0001832 | blastocyst growth | IEA | - | |
| GO:0006357 | regulation of transcription from RNA polymerase II promoter | TAS | 8208605 | |
| GO:0006350 | transcription | IEA | - | |
| GO:0006346 | methylation-dependent chromatin silencing | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000792 | heterochromatin | IEA | - | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005634 | nucleus | TAS | 8232556 | |
| GO:0005654 | nucleoplasm | TAS | 8208605 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTB | PS1TP5BP1 | actin, beta | - | HPRD,BioGRID | 9845365 |
| ACTL6A | ACTL6 | Arp4 | BAF53A | INO80K | MGC5382 | actin-like 6A | - | HPRD,BioGRID | 9845365 |
| AHR | - | aryl hydrocarbon receptor | - | HPRD,BioGRID | 11805098 |
| ALB | DKFZp779N1935 | PRO0883 | PRO0903 | PRO1341 | albumin | Brg-1 interactis with the Albumin gene. | BIND | 15616580 |
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | Affinity Capture-Western | BioGRID | 14966121 |
| ARID1A | B120 | BAF250 | BAF250a | BM029 | C1orf4 | P270 | SMARCF1 | AT rich interactive domain 1A (SWI-like) | Affinity Capture-Western Co-purification | BioGRID | 8895581 |9845365 |
| ARID1B | 6A3-5 | BAF250b | BRIGHT | DAN15 | ELD/OSA1 | KIAA1235 | p250R | AT rich interactive domain 1B (SWI1-like) | ELD/OSA1 interacts with BRG1. | BIND | 11988099 |
| ARID1B | 6A3-5 | BAF250b | BRIGHT | DAN15 | ELD/OSA1 | KIAA1235 | p250R | AT rich interactive domain 1B (SWI1-like) | - | HPRD,BioGRID | 11988099 |12200431 |
| BAZ1B | WBSCR10 | WBSCR9 | WSTF | bromodomain adjacent to zinc finger domain, 1B | - | HPRD | 12837248 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | - | HPRD,BioGRID | 10943845 |
| BRWD1 | C21orf107 | FLJ43918 | N143 | WDR9 | bromodomain and WD repeat domain containing 1 | - | HPRD | 12889071 |
| CBX5 | HP1 | HP1A | chromobox homolog 5 (HP1 alpha homolog, Drosophila) | - | HPRD,BioGRID | 12411497 |
| CCNE1 | CCNE | cyclin E1 | - | HPRD,BioGRID | 9891079 |
| CDKN1A | CAP20 | CDKN1 | CIP1 | MDA-6 | P21 | SDI1 | WAF1 | p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | BRG1 interacts with p21 promoter. | BIND | 15780937 |
| CEBPB | C/EBP-beta | CRP2 | IL6DBP | LAP | MGC32080 | NF-IL6 | TCF5 | CCAAT/enhancer binding protein (C/EBP), beta | - | HPRD | 10619021 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | Phenotypic Enhancement | BioGRID | 11003650 |
| CSF1 | MCSF | MGC31930 | colony stimulating factor 1 (macrophage) | BRG1 interacts with CSF1 promoter. | BIND | 15780937 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | - | HPRD,BioGRID | 11532957 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | - | HPRD,BioGRID | 9099865 |
| ETS2 | ETS2IT1 | v-ets erythroblastosis virus E26 oncogene homolog 2 (avian) | ETS2 (Ets-2) interacts with SMARCA4 (Brg-1). This interaction was modeled on a demonstrated interaction between ETS2 from an unspecified species and SMARCA4 from an unspecified species. | BIND | 12637547 |
| FANCA | FA | FA-H | FA1 | FAA | FACA | FAH | FANCH | MGC75158 | Fanconi anemia, complementation group A | - | HPRD,BioGRID | 11726552 |
| GATA1 | ERYF1 | GF-1 | GF1 | NFE1 | GATA binding protein 1 (globin transcription factor 1) | Reconstituted Complex | BioGRID | 11018012 |
| GRM2 | GLUR2 | GPRC1B | MGLUR2 | mGlu2 | glutamate receptor, metabotropic 2 | BRG1 interacts with mGluR2 promoter. | BIND | 15035981 |
| H3F3A | H3.3A | H3F3 | MGC87782 | MGC87783 | H3 histone, family 3A | H3F3A (Histone 3) interacts with SMARCA4 (BRG-1). | BIND | 15616580 |
| HDAC2 | RPD3 | YAF1 | histone deacetylase 2 | Co-purification | BioGRID | 11238380 |
| HDAC3 | HD3 | RPD3 | RPD3-2 | histone deacetylase 3 | - | HPRD | 10944117 |
| HNF1A | HNF1 | LFB1 | MODY3 | TCF1 | HNF1 homeobox A | Brg-1 interacts with the HNF-1 gene. | BIND | 15616580 |
| HNF4A | FLJ39654 | HNF4 | HNF4a7 | HNF4a8 | HNF4a9 | MODY | MODY1 | NR2A1 | NR2A21 | TCF | TCF14 | hepatocyte nuclear factor 4, alpha | Brg-1 interacts with the HNF-4 gene. | BIND | 15616580 |
| HSF1 | HSTF1 | heat shock transcription factor 1 | Affinity Capture-Western in vitro | BioGRID | 8856972 |11486022 |
| HSF1 | HSTF1 | heat shock transcription factor 1 | - | HPRD | 8856972 |9222609 |11486022 |
| HSP90B1 | ECGP | GP96 | GRP94 | TRA1 | heat shock protein 90kDa beta (Grp94), member 1 | Affinity Capture-Western | BioGRID | 11726552 |
| ING1 | p24ING1c | p33 | p33ING1 | p33ING1b | p47 | p47ING1a | inhibitor of growth family, member 1 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 11784859 |
| KLF1 | EKLF | Kruppel-like factor 1 (erythroid) | Reconstituted Complex | BioGRID | 11018012 |
| MED17 | CRSP6 | CRSP77 | DRIP80 | FLJ10812 | TRAP80 | mediator complex subunit 17 | Co-purification | BioGRID | 12154023 |
| MPHOSPH6 | MPP | MPP-6 | MPP6 | M-phase phosphoprotein 6 | - | HPRD | 15231747 |
| MPP6 | PALS2 | VAM-1 | VAM1 | p55T | membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) | - | HPRD,BioGRID | 15231747 |
| MYC | bHLHe39 | c-Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | Affinity Capture-MS Affinity Capture-Western | BioGRID | 11839798 |17353931 |
| NCOA1 | F-SRC-1 | KAT13A | MGC129719 | MGC129720 | NCoA-1 | RIP160 | SRC-1 | SRC1 | bHLHe42 | nuclear receptor coactivator 1 | Phenotypic Enhancement | BioGRID | 11003650 |
| NCOR1 | KIAA1047 | MGC104216 | N-CoR | TRAC1 | hCIT529I10 | hN-CoR | nuclear receptor co-repressor 1 | Co-purification | BioGRID | 11013263 |
| NCOR2 | CTG26 | SMRT | SMRTE | SMRTE-tau | TNRC14 | TRAC-1 | TRAC1 | nuclear receptor co-repressor 2 | Co-fractionation | BioGRID | 10944117 |
| NR3C1 | GCCR | GCR | GR | GRL | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | Affinity Capture-Western Reconstituted Complex | BioGRID | 10688647 |12917342 |
| PABPN1 | OPMD | PAB2 | PABP2 | poly(A) binding protein, nuclear 1 | Two-hybrid | BioGRID | 11371506 |
| PHB | PHB1 | prohibitin | - | HPRD,BioGRID | 12065415 |
| PHB | PHB1 | prohibitin | Prohibitin interacts with Brg-1 | BIND | 12065415 |
| POLR2A | MGC75453 | POLR2 | POLRA | RPB1 | RPBh1 | RPO2 | RPOL2 | RpIILS | hRPB220 | hsRPB1 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | Affinity Capture-Western Co-purification in vitro | BioGRID | 8895581 |9443979 |9845365 |11238380 |
| POLR2A | MGC75453 | POLR2 | POLRA | RPB1 | RPBh1 | RPO2 | RPOL2 | RpIILS | hRPB220 | hsRPB1 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | - | HPRD | 9443979 |12154023 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | - | HPRD,BioGRID | 7923370 |
| RBBP4 | NURF55 | RBAP48 | retinoblastoma binding protein 4 | Co-purification | BioGRID | 11238380 |
| RELB | I-REL | IREL | v-rel reticuloendotheliosis viral oncogene homolog B | Affinity Capture-MS | BioGRID | 14743216 |
| SIN3A | DKFZp434K2235 | FLJ90319 | KIAA0700 | SIN3 homolog A, transcription regulator (yeast) | Affinity Capture-Western Co-purification Reconstituted Complex | BioGRID | 11238380 |11784859 |
| SMARCB1 | BAF47 | INI1 | RDT | SNF5 | SNF5L1 | Sfh1p | Snr1 | hSNFS | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1 | - | HPRD,BioGRID | 8895581 |
| SMARCC1 | BAF155 | CRACC1 | Rsc8 | SRG3 | SWI3 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 | Affinity Capture-Western Co-purification | BioGRID | 8895581 |9845365 |11238380 |11726552 |
| SMARCC1 | BAF155 | CRACC1 | Rsc8 | SRG3 | SWI3 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 | - | HPRD | 9891079 |
| SMARCC2 | BAF170 | CRACC2 | Rsc8 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 | Affinity Capture-Western Co-purification | BioGRID | 8895581 |9845365 |11726552 |
| SMARCD1 | BAF60A | CRACD1 | Rsc6p | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 | Affinity Capture-Western | BioGRID | 12917342 |
| SMARCE1 | BAF57 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 | Affinity Capture-Western Co-purification | BioGRID | 8895581 |9845365 |
| SP1 | - | Sp1 transcription factor | Reconstituted Complex | BioGRID | 11018012 |
| SS18 | MGC116875 | SSXT | SYT | SYT-SSX1 | SYT-SSX2 | synovial sarcoma translocation, chromosome 18 | - | HPRD | 11734557|14603256 |
| STAT2 | ISGF-3 | MGC59816 | P113 | STAT113 | signal transducer and activator of transcription 2, 113kDa | - | HPRD,BioGRID | 12244326 |
| STK11 | LKB1 | PJS | serine/threonine kinase 11 | - | HPRD,BioGRID | 11445556 |
| STMN2 | SCG10 | SCGN10 | SGC10 | stathmin-like 2 | BRG1 interacts with SCG10 promoter. | BIND | 15035981 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | Affinity Capture-Western Co-fractionation Reconstituted Complex | BioGRID | 11950834 |
| VDR | NR1I1 | vitamin D (1,25- dihydroxyvitamin D3) receptor | Affinity Capture-Western | BioGRID | 12837248 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID REG GR PATHWAY | 82 | 60 | All SZGR 2.0 genes in this pathway |
| PID ERB GENOMIC PATHWAY | 15 | 8 | All SZGR 2.0 genes in this pathway |
| PID P53 DOWNSTREAM PATHWAY | 137 | 94 | All SZGR 2.0 genes in this pathway |
| PID BETA CATENIN NUC PATHWAY | 80 | 60 | All SZGR 2.0 genes in this pathway |
| PID RB 1PATHWAY | 65 | 46 | All SZGR 2.0 genes in this pathway |
| LIU SOX4 TARGETS DN | 309 | 191 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA PROGENITOR DN | 66 | 42 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY UP | 430 | 232 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED DN | 335 | 193 | All SZGR 2.0 genes in this pathway |
| HOEBEKE LYMPHOID STEM CELL UP | 95 | 64 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION MONOCYTE UP | 204 | 140 | All SZGR 2.0 genes in this pathway |
| HUMMEL BURKITTS LYMPHOMA UP | 43 | 27 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP UP | 265 | 158 | All SZGR 2.0 genes in this pathway |
| MAYBURD RESPONSE TO L663536 UP | 29 | 18 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 DN | 378 | 231 | All SZGR 2.0 genes in this pathway |
| NADERI BREAST CANCER PROGNOSIS UP | 50 | 26 | All SZGR 2.0 genes in this pathway |
| VANHARANTA UTERINE FIBROID WITH 7Q DELETION UP | 67 | 37 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 5 6WK DN | 137 | 97 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 6 7WK UP | 197 | 135 | All SZGR 2.0 genes in this pathway |
| MATTIOLI MGUS VS PCL | 116 | 62 | All SZGR 2.0 genes in this pathway |
| MATTIOLI MULTIPLE MYELOMA SUBGROUPS | 16 | 7 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| PUJANA BREAST CANCER LIT INT NETWORK | 101 | 73 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES CORE NINE CORRELATED | 100 | 68 | All SZGR 2.0 genes in this pathway |
| MORI LARGE PRE BII LYMPHOCYTE UP | 86 | 49 | All SZGR 2.0 genes in this pathway |
| MORI IMMATURE B LYMPHOCYTE DN | 90 | 55 | All SZGR 2.0 genes in this pathway |
| MORI MATURE B LYMPHOCYTE DN | 75 | 43 | All SZGR 2.0 genes in this pathway |
| MORI EMU MYC LYMPHOMA BY ONSET TIME UP | 110 | 69 | All SZGR 2.0 genes in this pathway |
| GOLUB ALL VS AML UP | 24 | 20 | All SZGR 2.0 genes in this pathway |
| CHANG POU5F1 TARGETS UP | 15 | 10 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST DN | 309 | 206 | All SZGR 2.0 genes in this pathway |
| GILDEA METASTASIS | 30 | 14 | All SZGR 2.0 genes in this pathway |
| MUNSHI MULTIPLE MYELOMA DN | 11 | 6 | All SZGR 2.0 genes in this pathway |
| HADDAD B LYMPHOCYTE PROGENITOR | 293 | 193 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI DN | 172 | 107 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR | 532 | 309 | All SZGR 2.0 genes in this pathway |
| RHODES CANCER META SIGNATURE | 64 | 47 | All SZGR 2.0 genes in this pathway |
| HENDRICKS SMARCA4 TARGETS UP | 56 | 35 | All SZGR 2.0 genes in this pathway |
| LEE AGING CEREBELLUM DN | 86 | 66 | All SZGR 2.0 genes in this pathway |
| MEDINA SMARCA4 TARGETS | 44 | 29 | All SZGR 2.0 genes in this pathway |
| MCCLUNG COCAIN REWARD 4WK | 75 | 47 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS DN | 141 | 92 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE UP | 393 | 244 | All SZGR 2.0 genes in this pathway |
| HOWLIN CITED1 TARGETS 2 UP | 17 | 10 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR UP | 294 | 199 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| HOFFMANN SMALL PRE BII TO IMMATURE B LYMPHOCYTE DN | 50 | 33 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A5 | 70 | 32 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR DN | 277 | 166 | All SZGR 2.0 genes in this pathway |
| FOURNIER ACINAR DEVELOPMENT LATE 2 | 277 | 172 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA UP | 207 | 143 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 14 | 143 | 86 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO GONADOTROPHINS DN | 87 | 66 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO FORSKOLIN UP | 90 | 58 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| BILANGES SERUM SENSITIVE VIA TSC2 | 39 | 25 | All SZGR 2.0 genes in this pathway |
| PECE MAMMARY STEM CELL UP | 146 | 75 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-1/206 | 7 | 14 | 1A,m8 | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-101 | 358 | 364 | 1A | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-139 | 313 | 319 | m8 | hsa-miR-139brain | UCUACAGUGCACGUGUCU |
| miR-144 | 358 | 364 | 1A | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-155 | 204 | 211 | 1A,m8 | hsa-miR-155 | UUAAUGCUAAUCGUGAUAGGGG |
| miR-199 | 417 | 423 | 1A | hsa-miR-199a | CCCAGUGUUCAGACUACCUGUUC |
| hsa-miR-199b | CCCAGUGUUUAGACUAUCUGUUC | ||||
| miR-324-5p | 328 | 334 | 1A | hsa-miR-324-5p | CGCAUCCCCUAGGGCAUUGGUGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.