Gene Page: TP53
Summary ?
| GeneID | 7157 |
| Symbol | TP53 |
| Synonyms | BCC7|LFS1|P53|TRP53 |
| Description | tumor protein p53 |
| Reference | MIM:191170|HGNC:HGNC:11998|Ensembl:ENSG00000141510|HPRD:01859|Vega:OTTHUMG00000162125 |
| Gene type | protein-coding |
| Map location | 17p13.1 |
| Pascal p-value | 0.206 |
| Fetal beta | 0.452 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 2 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 3 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 31.3933 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
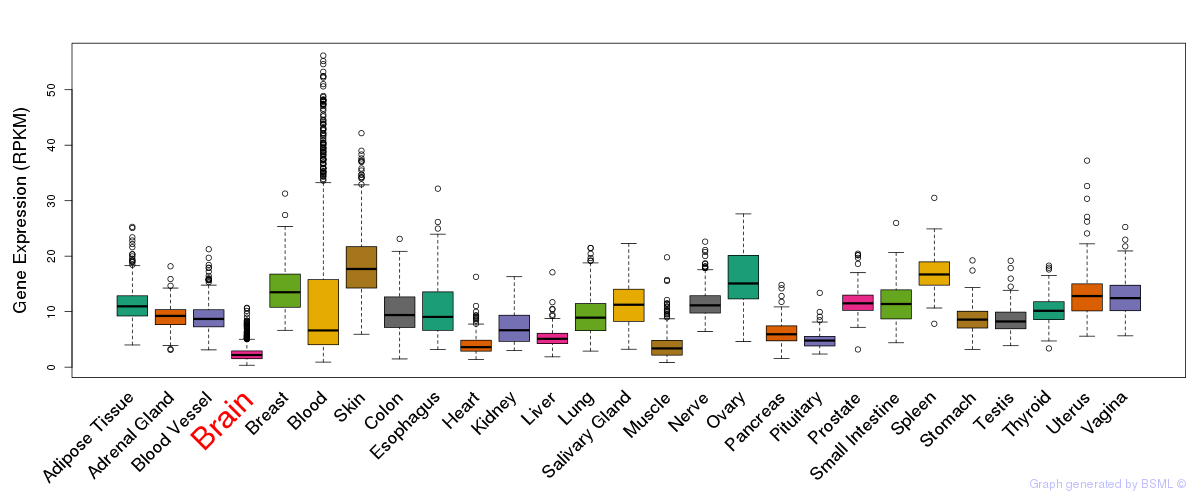
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ANO2 | 0.86 | 0.28 |
| LHX9 | 0.83 | 0.44 |
| EPHA1 | 0.83 | 0.19 |
| KLK10 | 0.79 | -0.01 |
| FOXP2 | 0.78 | 0.45 |
| TRPM8 | 0.77 | 0.09 |
| QRFPR | 0.75 | -0.07 |
| MAP4K1 | 0.75 | 0.28 |
| VAT1 | 0.75 | 0.79 |
| SLC26A11 | 0.75 | 0.55 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.30 | -0.72 |
| CA4 | -0.28 | -0.69 |
| PTGDS | -0.28 | -0.68 |
| HLA-F | -0.28 | -0.69 |
| SPARCL1 | -0.28 | -0.64 |
| CCNI2 | -0.28 | -0.64 |
| TESK2 | -0.28 | -0.67 |
| TINAGL1 | -0.27 | -0.60 |
| AF347015.27 | -0.27 | -0.71 |
| ALDOC | -0.27 | -0.65 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000739 | DNA strand annealing activity | IDA | 8183576 | |
| GO:0003682 | chromatin binding | IDA | 17599062 | |
| GO:0003700 | transcription factor activity | IDA | 7587074 | |
| GO:0003700 | transcription factor activity | IEA | - | |
| GO:0005507 | copper ion binding | IDA | 7824276 | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 15782130 | |
| GO:0005524 | ATP binding | IDA | 8183576 | |
| GO:0004518 | nuclease activity | TAS | 11002423 | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0008270 | zinc ion binding | TAS | 10065153 | |
| GO:0051087 | chaperone binding | IPI | 18086682 | |
| GO:0019899 | enzyme binding | IPI | 10666337 |15577914 | |
| GO:0046872 | metal ion binding | IEA | - | |
| GO:0047485 | protein N-terminus binding | IPI | 11861836 | |
| GO:0046982 | protein heterodimerization activity | IPI | 10837489 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007417 | central nervous system development | IEA | Brain (GO term level: 6) | - |
| GO:0007406 | negative regulation of neuroblast proliferation | IEA | neurogenesis (GO term level: 10) | - |
| GO:0043525 | positive regulation of neuron apoptosis | IEA | neuron (GO term level: 9) | - |
| GO:0000060 | protein import into nucleus, translocation | IEA | - | |
| GO:0001701 | in utero embryonic development | IEA | - | |
| GO:0002347 | response to tumor cell | IEA | - | |
| GO:0001836 | release of cytochrome c from mitochondria | IEA | - | |
| GO:0006355 | regulation of transcription, DNA-dependent | IDA | 7587074 | |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006461 | protein complex assembly | IDA | 12915590 | |
| GO:0006289 | nucleotide-excision repair | IMP | 7663514 | |
| GO:0006284 | base-excision repair | TAS | 15116721 | |
| GO:0006917 | induction of apoptosis | IEA | - | |
| GO:0007050 | cell cycle arrest | TAS | 8675009 | |
| GO:0009792 | embryonic development ending in birth or egg hatching | IEA | - | |
| GO:0008283 | cell proliferation | TAS | 10065150 | |
| GO:0008629 | induction of apoptosis by intracellular signals | TAS | 16462759 | |
| GO:0008635 | caspase activation via cytochrome c | IDA | 12667443 | |
| GO:0010165 | response to X-ray | IEA | - | |
| GO:0010332 | response to gamma radiation | IEA | - | |
| GO:0008156 | negative regulation of DNA replication | IEA | - | |
| GO:0008104 | protein localization | IDA | 16507995 | |
| GO:0007569 | cell aging | IMP | 12080348 | |
| GO:0009411 | response to UV | IEA | - | |
| GO:0006983 | ER overload response | IDA | 14744935 | |
| GO:0006974 | response to DNA damage stimulus | IEA | - | |
| GO:0006974 | response to DNA damage stimulus | IEP | 8986812 | |
| GO:0006915 | apoptosis | IEA | - | |
| GO:0007275 | multicellular organismal development | IMP | 10065150 | |
| GO:0043066 | negative regulation of apoptosis | IEA | - | |
| GO:0018144 | RNA-protein covalent cross-linking | IEA | - | |
| GO:0030154 | cell differentiation | TAS | 10065150 | |
| GO:0051097 | negative regulation of helicase activity | TAS | 7663514 | |
| GO:0042149 | cellular response to glucose starvation | IDA | 14744935 | |
| GO:0042771 | DNA damage response, signal transduction by p53 class mediator resulting in induction of apoptosis | IDA | 14654789 |14744935 | |
| GO:0042771 | DNA damage response, signal transduction by p53 class mediator resulting in induction of apoptosis | IEA | - | |
| GO:0051262 | protein tetramerization | TAS | 15116721 | |
| GO:0031571 | G1 DNA damage checkpoint | IEA | - | |
| GO:0030308 | negative regulation of cell growth | IEA | - | |
| GO:0030308 | negative regulation of cell growth | IMP | 8986812 | |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | IDA | 17599062 | |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | IEA | - | |
| GO:0046902 | regulation of mitochondrial membrane permeability | TAS | 15116721 | |
| GO:0048147 | negative regulation of fibroblast proliferation | IEA | - | |
| GO:0045786 | negative regulation of cell cycle | IEA | - | |
| GO:0044419 | interspecies interaction between organisms | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | IEA | - | |
| GO:0005626 | insoluble fraction | IDA | 12915590 | |
| GO:0005634 | nucleus | IDA | 7720704 |16507995 | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005654 | nucleoplasm | IDA | 11080164 |12915590 | |
| GO:0005657 | replication fork | IEA | - | |
| GO:0005730 | nucleolus | IDA | 12080348 | |
| GO:0005737 | cytoplasm | IDA | 7720704 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005739 | mitochondrion | IDA | 12667443 | |
| GO:0005783 | endoplasmic reticulum | IEA | - | |
| GO:0016363 | nuclear matrix | IDA | 11080164 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABL1 | ABL | JTK7 | bcr/abl | c-ABL | p150 | v-abl | c-abl oncogene 1, receptor tyrosine kinase | - | HPRD,BioGRID | 10629029 |10713716 |
| ANKRD2 | ARPP | MGC104314 | ankyrin repeat domain 2 (stretch responsive muscle) | - | HPRD,BioGRID | 15136035 |
| ANXA3 | ANX3 | annexin A3 | Two-hybrid | BioGRID | 16169070 |
| APEX1 | APE | APE-1 | APE1 | APEN | APEX | APX | HAP1 | REF-1 | REF1 | APEX nuclease (multifunctional DNA repair enzyme) 1 | p53 interacts with Ref-1. | BIND | 15824742 |
| APEX1 | APE | APE-1 | APE1 | APEN | APEX | APX | HAP1 | REF-1 | REF1 | APEX nuclease (multifunctional DNA repair enzyme) 1 | - | HPRD,BioGRID | 9119221 |
| APTX | AOA | AOA1 | AXA1 | EAOH | EOAHA | FHA-HIT | FLJ20157 | MGC1072 | aprataxin | Affinity Capture-Western Reconstituted Complex | BioGRID | 15044383 |
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | Biochemical Activity Phenotypic Suppression | BioGRID | 11504717 |
| ARID3A | BRIGHT | DRIL1 | DRIL3 | E2FBP1 | AT rich interactive domain 3A (BRIGHT-like) | - | HPRD | 12136662 |
| ARIH2 | ARI2 | FLJ10938 | FLJ33921 | TRIAD1 | ariadne homolog 2 (Drosophila) | Two-hybrid | BioGRID | 16169070 |
| ARL3 | ARFL3 | ADP-ribosylation factor-like 3 | Two-hybrid | BioGRID | 16169070 |
| ATF3 | - | activating transcription factor 3 | - | HPRD,BioGRID | 11792711 |
| ATF3 | - | activating transcription factor 3 | An unspecified isoform of ATF3 interacts with p53. | BIND | 15933712 |
| ATM | AT1 | ATA | ATC | ATD | ATDC | ATE | DKFZp781A0353 | MGC74674 | TEL1 | TELO1 | ataxia telangiectasia mutated | - | HPRD,BioGRID | 9843217 |
| ATM | AT1 | ATA | ATC | ATD | ATDC | ATE | DKFZp781A0353 | MGC74674 | TEL1 | TELO1 | ataxia telangiectasia mutated | ATM phosphorylates p53 at serine 15. | BIND | 15064416 |
| ATM | AT1 | ATA | ATC | ATD | ATDC | ATE | DKFZp781A0353 | MGC74674 | TEL1 | TELO1 | ataxia telangiectasia mutated | An unspecified isoform of ATM interacts with and phosphorylates p53. This interaction was modeled on a demonstrated interaction between human ATM and p53 from an unspecified species. | BIND | 15916964 |
| ATM | AT1 | ATA | ATC | ATD | ATDC | ATE | DKFZp781A0353 | MGC74674 | TEL1 | TELO1 | ataxia telangiectasia mutated | ATM phosphorylates p53 at Ser15. | BIND | 15175154 |
| ATM | AT1 | ATA | ATC | ATD | ATDC | ATE | DKFZp781A0353 | MGC74674 | TEL1 | TELO1 | ataxia telangiectasia mutated | ATM phosphorylates p53 at serine 15. | BIND | 15790808 |
| ATR | FRP1 | MEC1 | SCKL | SCKL1 | ataxia telangiectasia and Rad3 related | ATR interacts with TP53 (p53). | BIND | 15775976 |
| ATR | FRP1 | MEC1 | SCKL | SCKL1 | ataxia telangiectasia and Rad3 related | Protein-peptide Reconstituted Complex | BioGRID | 10608806 |15159397 |
| AURKA | AIK | ARK1 | AURA | AURORA2 | BTAK | MGC34538 | STK15 | STK6 | STK7 | aurora kinase A | p53 interacts with STK15. | BIND | 12198151 |
| AURKA | AIK | ARK1 | AURA | AURORA2 | BTAK | MGC34538 | STK15 | STK6 | STK7 | aurora kinase A | - | HPRD,BioGRID | 12198151 |
| BAG1 | RAP46 | BCL2-associated athanogene | Phenotypic Suppression | BioGRID | 9582267 |
| BAK1 | BAK | BAK-LIKE | BCL2L7 | CDN1 | MGC117255 | MGC3887 | BCL2-antagonist/killer 1 | Mitochondrial p53-P72 interacts with Bak. | BIND | 15077116 |
| BAK1 | BAK | BAK-LIKE | BCL2L7 | CDN1 | MGC117255 | MGC3887 | BCL2-antagonist/killer 1 | Mitochondrial p53-R72 interacts with Bak. | BIND | 15077116 |
| BAK1 | BAK | BAK-LIKE | BCL2L7 | CDN1 | MGC117255 | MGC3887 | BCL2-antagonist/killer 1 | Affinity Capture-MS Affinity Capture-Western Reconstituted Complex | BioGRID | 15077116 |
| BANP | BEND1 | DKFZp761H172 | FLJ10177 | FLJ20538 | SMAR1 | SMARBP1 | BTG3 associated nuclear protein | - | HPRD | 15701641 |
| BARD1 | - | BRCA1 associated RING domain 1 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 14636569 |
| BARD1 | - | BRCA1 associated RING domain 1 | TP53 (p53) interacts with BARD1. | BIND | 15782130 |
| BAX | BCL2L4 | BCL2-associated X protein | p53 interacts with Bax. | BIND | 15607964 |
| BAX | BCL2L4 | BCL2-associated X protein | p53 interacts with bax promoter. | BIND | 16009130 |
| BAX | BCL2L4 | BCL2-associated X protein | p53 interacts with the Bax promoter. | BIND | 15710329 |
| BCL2 | Bcl-2 | B-cell CLL/lymphoma 2 | - | HPRD | 12667443 |
| BCL2L1 | BCL-XL/S | BCL2L | BCLX | Bcl-X | DKFZp781P2092 | bcl-xL | bcl-xS | BCL2-like 1 | Bcl-xl interacts with p53. | BIND | 14963330 |
| BCL2L1 | BCL-XL/S | BCL2L | BCLX | Bcl-X | DKFZp781P2092 | bcl-xL | bcl-xS | BCL2-like 1 | - | HPRD | 12667443 |
| BCL6 | BCL5 | BCL6A | LAZ3 | ZBTB27 | ZNF51 | B-cell CLL/lymphoma 6 | BCL6 interacts with the p53 promoter region. | BIND | 15577913 |
| BCR | ALL | BCR-ABL1 | BCR1 | CML | D22S11 | D22S662 | FLJ16453 | PHL | breakpoint cluster region | Two-hybrid | BioGRID | 16169070 |
| BLM | BS | MGC126616 | MGC131618 | MGC131620 | RECQ2 | RECQL2 | RECQL3 | Bloom syndrome | BLM interacts with p53. | BIND | 12080066 |
| BLM | BS | MGC126616 | MGC131618 | MGC131620 | RECQ2 | RECQL2 | RECQL3 | Bloom syndrome | BLM interacts with p53. This interaction was modeled on a demonstrated interaction between BLM and p53 both from African green monkey. | BIND | 15806145 |
| BLM | BS | MGC126616 | MGC131618 | MGC131620 | RECQ2 | RECQL2 | RECQL3 | Bloom syndrome | - | HPRD,BioGRID | 11781842 |
| BLM | BS | MGC126616 | MGC131618 | MGC131620 | RECQ2 | RECQL2 | RECQL3 | Bloom syndrome | p53 interacts with BLM. | BIND | 11781842 |
| BNIP3L | BNIP3a | NIX | BCL2/adenovirus E1B 19kDa interacting protein 3-like | p53 interacts with Bnip3L. | BIND | 15607964 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | BRCA1a interacts with p53. | BIND | 9926942 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | BRCA1 physically associates with p53 and stimulates its transcriptional activity | BIND | 9582019 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | - | HPRD,BioGRID | 9482880 |
| BRCA2 | BRCC2 | FACD | FAD | FAD1 | FANCB | FANCD | FANCD1 | breast cancer 2, early onset | - | HPRD,BioGRID | 9811893 |
| BRCC3 | BRCC36 | C6.1A | CXorf53 | BRCA1/BRCA2-containing complex, subunit 3 | Affinity Capture-MS Affinity Capture-Western Co-purification | BioGRID | 14636569 |
| BRE | BRCC4 | BRCC45 | brain and reproductive organ-expressed (TNFRSF1A modulator) | Affinity Capture-MS Affinity Capture-Western Co-purification | BioGRID | 14636569 |
| BRF1 | BRF | FLJ42674 | FLJ43034 | GTF3B | MGC105048 | TAF3B2 | TAF3C | TAFIII90 | TF3B90 | TFIIIB90 | hBRF | BRF1 homolog, subunit of RNA polymerase III transcription initiation factor IIIB (S. cerevisiae) | - | HPRD,BioGRID | 8943363 |
| BTBD2 | - | BTB (POZ) domain containing 2 | Two-hybrid | BioGRID | 16169070 |
| CABLES1 | CABLES | FLJ35924 | HsT2563 | IK3-1 | Cdk5 and Abl enzyme substrate 1 | - | HPRD,BioGRID | 14637168 |
| CABLES1 | CABLES | FLJ35924 | HsT2563 | IK3-1 | Cdk5 and Abl enzyme substrate 1 | - | HPRD | 11706030 |
| CABLES2 | C20orf150 | dJ908M14.2 | ik3-2 | Cdk5 and Abl enzyme substrate 2 | - | HPRD,BioGRID | 14637168 |
| CARM1 | PRMT4 | coactivator-associated arginine methyltransferase 1 | Reconstituted Complex | BioGRID | 15186775 |
| CARM1 | PRMT4 | coactivator-associated arginine methyltransferase 1 | CARM1 interacts with p53. This interaction was modeled on a demonstrated interaction between mouse CARM1 and human p53. | BIND | 15186775 |
| CCDC106 | HSU79303 | ZNF581 | coiled-coil domain containing 106 | Two-hybrid | BioGRID | 16169070 |
| CCL18 | AMAC-1 | AMAC1 | CKb7 | DC-CK1 | DCCK1 | MIP-4 | PARC | SCYA18 | chemokine (C-C motif) ligand 18 (pulmonary and activation-regulated) | Two-hybrid | BioGRID | 16169070 |
| CCNA2 | CCN1 | CCNA | cyclin A2 | - | HPRD,BioGRID | 10884347 |
| CCNG1 | CCNG | cyclin G1 | - | HPRD,BioGRID | 12556559 |
| CCNH | CAK | p34 | p37 | cyclin H | Affinity Capture-Western Far Western | BioGRID | 9840937 |
| CCT5 | CCT-epsilon | CCTE | KIAA0098 | TCP-1-epsilon | chaperonin containing TCP1, subunit 5 (epsilon) | Two-hybrid | BioGRID | 16169070 |
| CDC14A | cdc14 | hCDC14 | CDC14 cell division cycle 14 homolog A (S. cerevisiae) | - | HPRD,BioGRID | 10644693 |
| CDC14B | CDC14B3 | Cdc14B1 | Cdc14B2 | hCDC14B | CDC14 cell division cycle 14 homolog B (S. cerevisiae) | - | HPRD,BioGRID | 10644693 |
| CDC2 | CDC28A | CDK1 | DKFZp686L20222 | MGC111195 | cell division cycle 2, G1 to S and G2 to M | - | HPRD,BioGRID | 11327730 |
| CDC25C | CDC25 | cell division cycle 25 homolog C (S. pombe) | - | HPRD | 10853038 |
| CDC25C | CDC25 | cell division cycle 25 homolog C (S. pombe) | p53 interacts with the cdc25c promoter. | BIND | 15574328 |
| CDC42 | CDC42Hs | G25K | cell division cycle 42 (GTP binding protein, 25kDa) | Two-hybrid | BioGRID | 16169070 |
| CDK2 | p33(CDK2) | cyclin-dependent kinase 2 | Biochemical Activity | BioGRID | 10884347 |
| CDK7 | CAK1 | CDKN7 | MO15 | STK1 | p39MO15 | cyclin-dependent kinase 7 | Affinity Capture-Western Biochemical Activity | BioGRID | 9372954 |9840937 |
| CDK9 | C-2k | CDC2L4 | CTK1 | PITALRE | TAK | cyclin-dependent kinase 9 | Phenotypic Suppression | BioGRID | 10656684 |
| CDKN1A | CAP20 | CDKN1 | CIP1 | MDA-6 | P21 | SDI1 | WAF1 | p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | p53 interacts with p21 promoter. | BIND | 16009130 |
| CDKN1A | CAP20 | CDKN1 | CIP1 | MDA-6 | P21 | SDI1 | WAF1 | p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | TP53 (p53) interacts with the CDKN1A (p21) promoter. | BIND | 15674334 |
| CDKN1A | CAP20 | CDKN1 | CIP1 | MDA-6 | P21 | SDI1 | WAF1 | p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | p53 interacts with the p21 promoter. | BIND | 15660129 |
| CDKN1A | CAP20 | CDKN1 | CIP1 | MDA-6 | P21 | SDI1 | WAF1 | p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | Affinity Capture-Western | BioGRID | 11896572 |
| CDKN1A | CAP20 | CDKN1 | CIP1 | MDA-6 | P21 | SDI1 | WAF1 | p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | p53 interacts with p21 promoter. | BIND | 15989967 |
| CDKN1A | CAP20 | CDKN1 | CIP1 | MDA-6 | P21 | SDI1 | WAF1 | p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | p53 interacts with the p21 promoter. | BIND | 15710329 |
| CDKN2A | ARF | CDK4I | CDKN2 | CMM2 | INK4 | INK4a | MLM | MTS1 | TP16 | p14 | p14ARF | p16 | p16INK4 | p16INK4a | p19 | cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | Affinity Capture-Western | BioGRID | 9529249 |12446718 |14612427 |
| CDKN2C | INK4C | p18 | p18-INK4C | cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) | Two-hybrid | BioGRID | 16169070 |
| CEBPZ | CBF2 | HSP-CBF | NOC1 | CCAAT/enhancer binding protein (C/EBP), zeta | - | HPRD,BioGRID | 12534345 |
| CHD3 | Mi-2a | Mi2-ALPHA | ZFH | chromodomain helicase DNA binding protein 3 | - | HPRD,BioGRID | 10961991 |
| CHEK1 | CHK1 | CHK1 checkpoint homolog (S. pombe) | - | HPRD,BioGRID | 11896572 |
| CHUK | IKBKA | IKK-alpha | IKK1 | IKKA | NFKBIKA | TCF16 | conserved helix-loop-helix ubiquitous kinase | - | HPRD | 11805286 |
| COX17 | MGC104397 | MGC117386 | COX17 cytochrome c oxidase assembly homolog (S. cerevisiae) | Two-hybrid | BioGRID | 16169070 |
| CR2 | C3DR | CD21 | SLEB9 | complement component (3d/Epstein Barr virus) receptor 2 | P53 interacts with CR2. | BIND | 7753047 |
| CR2 | C3DR | CD21 | SLEB9 | complement component (3d/Epstein Barr virus) receptor 2 | - | HPRD,BioGRID | 7753047 |
| CREB1 | CREB | MGC9284 | cAMP responsive element binding protein 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 10848610 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | - | HPRD,BioGRID | 10848610 |11782467 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | p53 interacts with CBP. This interaction was modeled on a demonstrated interaction between human p53 and CBP from an unspecified species. | BIND | 10823891 |
| CSNK1D | HCKID | casein kinase 1, delta | - | HPRD | 9349507 |
| CSNK1E | HCKIE | MGC10398 | casein kinase 1, epsilon | - | HPRD | 9349507 |
| CSNK2B | CK2B | CK2N | CSK2B | G5A | MGC138222 | MGC138224 | casein kinase 2, beta polypeptide | - | HPRD | 10214938 |
| DDX5 | DKFZp686J01190 | G17P1 | HLR1 | HUMP68 | p68 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 | p68 interacts with p53. | BIND | 15660129 |
| DHCR24 | KIAA0018 | Nbla03646 | SELADIN1 | seladin-1 | 24-dehydrocholesterol reductase | Seladin-1 interacts with p53. | BIND | 15577914 |
| DLEU1 | BCMS | DLB1 | DLEU2 | LEU1 | LEU2 | MGC22430 | NCRNA00021 | XTP6 | deleted in lymphocytic leukemia 1 (non-protein coding) | Two-hybrid | BioGRID | 16169070 |
| DVL2 | - | dishevelled, dsh homolog 2 (Drosophila) | Two-hybrid | BioGRID | 16189514 |
| E4F1 | E4F | MGC99614 | E4F transcription factor 1 | - | HPRD,BioGRID | 10644996 |12446718|12446718 |
| EEF2 | EEF-2 | EF2 | eukaryotic translation elongation factor 2 | - | HPRD,BioGRID | 12891704 |
| EFEMP2 | FBLN4 | MBP1 | UPH1 | EGF-containing fibulin-like extracellular matrix protein 2 | - | HPRD,BioGRID | 10380882 |
| EGR1 | AT225 | G0S30 | KROX-24 | NGFI-A | TIS8 | ZIF-268 | ZNF225 | early growth response 1 | - | HPRD,BioGRID | 11251186 |
| EIF2AK2 | EIF2AK1 | MGC126524 | PKR | PRKR | eukaryotic translation initiation factor 2-alpha kinase 2 | - | HPRD,BioGRID | 10348343 |
| EIF2S2 | DKFZp686L18198 | EIF2 | EIF2B | EIF2beta | MGC8508 | eukaryotic translation initiation factor 2, subunit 2 beta, 38kDa | Two-hybrid | BioGRID | 16169070 |
| ELL | C19orf17 | DKFZp434I1916 | ELL1 | Men | elongation factor RNA polymerase II | - | HPRD,BioGRID | 10358050 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | p300 interacts with p53 | BIND | 15558054 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | p300 interacts with p53. This interaction was modeled on a demonstrated interaction between p300 from an unspecified species and human p53. | BIND | 15186775 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | p300 interacts with and acetylates p53. This interaction was modeled on a demonstrated interaction between p300 from an unspecified species and human p53. | BIND | 15933712 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | - | HPRD,BioGRID | 10942770 |
| EPHA3 | ETK | ETK1 | HEK | HEK4 | TYRO4 | EPH receptor A3 | Reconstituted Complex | BioGRID | 15355990 |
| ERCC2 | COFS2 | EM9 | MGC102762 | MGC126218 | MGC126219 | TTD | XPD | excision repair cross-complementing rodent repair deficiency, complementation group 2 | - | HPRD,BioGRID | 7663514 |
| ERCC3 | BTF2 | GTF2H | RAD25 | TFIIH | XPB | excision repair cross-complementing rodent repair deficiency, complementation group 3 (xeroderma pigmentosum group B complementing) | p53 interacts with ERCC3. This interaction was modeled on a demonstrated interaction between human p53 and ERCC3 from an unspecified species. | BIND | 12379483 |
| ERCC3 | BTF2 | GTF2H | RAD25 | TFIIH | XPB | excision repair cross-complementing rodent repair deficiency, complementation group 3 (xeroderma pigmentosum group B complementing) | - | HPRD,BioGRID | 7663514 |
| ERCC6 | ARMD5 | CKN2 | COFS | COFS1 | CSB | RAD26 | excision repair cross-complementing rodent repair deficiency, complementation group 6 | - | HPRD,BioGRID | 10882116 |
| ERH | DROER | FLJ27340 | enhancer of rudimentary homolog (Drosophila) | Two-hybrid | BioGRID | 16169070 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | - | HPRD,BioGRID | 10766163 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | P53 interacts with ER-alpha. | BIND | 10766163 |
| FAM173A | C16orf24 | MGC2494 | family with sequence similarity 173, member A | Two-hybrid | BioGRID | 16169070 |
| FOXO3 | AF6q21 | DKFZp781A0677 | FKHRL1 | FKHRL1P2 | FOXO2 | FOXO3A | MGC12739 | MGC31925 | forkhead box O3 | Foxo3a interacts with p53. | BIND | 15604409 |
| FXYD6 | - | FXYD domain containing ion transport regulator 6 | Two-hybrid | BioGRID | 16169070 |
| GNL3 | C77032 | E2IG3 | MGC800 | NS | guanine nucleotide binding protein-like 3 (nucleolar) | - | HPRD,BioGRID | 12464630 |
| GPS2 | AMF-1 | MGC104294 | MGC119287 | MGC119288 | MGC119289 | G protein pathway suppressor 2 | - | HPRD,BioGRID | 11486030 |
| GPS2 | AMF-1 | MGC104294 | MGC119287 | MGC119288 | MGC119289 | G protein pathway suppressor 2 | p53 interacts with AMF1 | BIND | 11486030 |
| GSK3B | - | glycogen synthase kinase 3 beta | p53 interacts with GSK3-beta. | BIND | 12048243 |
| GSK3B | - | glycogen synthase kinase 3 beta | - | HPRD,BioGRID | 12048243 |
| GSTM4 | GSTM4-4 | GTM4 | MGC131945 | MGC9247 | glutathione S-transferase mu 4 | Two-hybrid | BioGRID | 16169070 |
| GTF2H1 | BTF2 | TFB1 | TFIIH | general transcription factor IIH, polypeptide 1, 62kDa | - | HPRD,BioGRID | 7935417 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | Affinity Capture-Western | BioGRID | 12426395 |
| HIF1A | HIF-1alpha | HIF1 | HIF1-ALPHA | MOP1 | PASD8 | hypoxia-inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) | Affinity Capture-Western Reconstituted Complex | BioGRID | 9537326 |10640274 |12124396 |12606552 |
| HIF1A | HIF-1alpha | HIF1 | HIF1-ALPHA | MOP1 | PASD8 | hypoxia-inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) | - | HPRD | 9537326|12124396 |
| HIF1A | HIF-1alpha | HIF1 | HIF1-ALPHA | MOP1 | PASD8 | hypoxia-inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) | p53 interacts with HIF-1 alpha. | BIND | 15629713 |
| HIF1A | HIF-1alpha | HIF1 | HIF1-ALPHA | MOP1 | PASD8 | hypoxia-inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) | HIF-1alpha interacts with p53 | BIND | 11593383 |
| HIPK1 | KIAA0630 | MGC26642 | MGC33446 | MGC33548 | Myak | Nbak2 | homeodomain interacting protein kinase 1 | - | HPRD,BioGRID | 12702766 |
| HIPK2 | DKFZp686K02111 | FLJ23711 | PRO0593 | homeodomain interacting protein kinase 2 | Affinity Capture-Western Biochemical Activity Reconstituted Complex Two-hybrid | BioGRID | 11740489 |11925430 |
| HIPK2 | DKFZp686K02111 | FLJ23711 | PRO0593 | homeodomain interacting protein kinase 2 | HIPK2 does not interact directly with tumour suppressor protein p53 but enhances p53 activity and p53 expression or stability | BIND | 11532197 |
| HIPK3 | DYRK6 | FIST3 | PKY | YAK1 | homeodomain interacting protein kinase 3 | Two-hybrid | BioGRID | 10961991 |
| HMGB1 | DKFZp686A04236 | HMG1 | HMG3 | SBP-1 | high-mobility group box 1 | p53 interacts with HMG-1. | BIND | 9472015 |
| HMGB1 | DKFZp686A04236 | HMG1 | HMG3 | SBP-1 | high-mobility group box 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 11106654 |11748221 |
| HMGB1 | DKFZp686A04236 | HMG1 | HMG3 | SBP-1 | high-mobility group box 1 | - | HPRD | 9472015 |
| HMGB2 | HMG2 | high-mobility group box 2 | - | HPRD | 11748232 |
| HNF4A | FLJ39654 | HNF4 | HNF4a7 | HNF4a8 | HNF4a9 | MODY | MODY1 | NR2A1 | NR2A21 | TCF | TCF14 | hepatocyte nuclear factor 4, alpha | - | HPRD,BioGRID | 11818510 |
| HSP90AA1 | FLJ31884 | HSP86 | HSP89A | HSP90A | HSP90N | HSPC1 | HSPCA | HSPCAL1 | HSPCAL4 | HSPN | Hsp89 | Hsp90 | LAP2 | heat shock protein 90kDa alpha (cytosolic), class A member 1 | - | HPRD,BioGRID | 11507088 |12427754 |
| HSPA1A | FLJ54303 | FLJ54370 | FLJ54392 | FLJ54408 | FLJ75127 | HSP70-1 | HSP70-1A | HSP70I | HSP72 | HSPA1 | HSPA1B | heat shock 70kDa protein 1A | - | HPRD | 7811761 |
| HSPA8 | HSC54 | HSC70 | HSC71 | HSP71 | HSP73 | HSPA10 | LAP1 | MGC131511 | MGC29929 | NIP71 | heat shock 70kDa protein 8 | Hsc70 interacts with p53. | BIND | 8940078 |
| HSPA8 | HSC54 | HSC70 | HSC71 | HSP71 | HSP73 | HSPA10 | LAP1 | MGC131511 | MGC29929 | NIP71 | heat shock 70kDa protein 8 | Reconstituted Complex | BioGRID | 11297531 |
| HSPA8 | HSC54 | HSC70 | HSC71 | HSP71 | HSP73 | HSPA10 | LAP1 | MGC131511 | MGC29929 | NIP71 | heat shock 70kDa protein 8 | p53 interacts with an unspecified isoform of Hsc70. This interaction was modeled on a demonstrated interaction between human p53 and bovine Hsc70. | BIND | 9235949 |
| HSPA9 | CSA | GRP75 | HSPA9B | MGC4500 | MOT | MOT2 | MTHSP75 | PBP74 | mot-2 | heat shock 70kDa protein 9 (mortalin) | - | HPRD,BioGRID | 11900485 |
| HSPB1 | CMT2F | DKFZp586P1322 | HMN2B | HS.76067 | HSP27 | HSP28 | Hsp25 | SRP27 | heat shock 27kDa protein 1 | Two-hybrid | BioGRID | 16169070 |
| HTT | HD | IT15 | huntingtin | p53 interacts with htt. This interaction was modeled on a demonstrated interaction between p53 from human and htt from an unspecified species. | BIND | 10823891 |
| HTT | HD | IT15 | huntingtin | Htt interacts with TP53(p53). | BIND | 15996546 |
| HTT | HD | IT15 | huntingtin | - | HPRD,BioGRID | 10823891 |
| HUWE1 | ARF-BP1 | HECTH9 | HSPC272 | Ib772 | KIAA0312 | LASU1 | MULE | UREB1 | HECT, UBA and WWE domain containing 1 | ARF-BP1 interacts with and ubiquitinates p53. | BIND | 15989956 |
| IFI16 | IFNGIP1 | MGC9466 | PYHIN2 | interferon, gamma-inducible protein 16 | - | HPRD,BioGRID | 11146555 |
| ING1 | p24ING1c | p33 | p33ING1 | p33ING1b | p47 | p47ING1a | inhibitor of growth family, member 1 | - | HPRD,BioGRID | 9440695 |12208736 |
| ING1 | p24ING1c | p33 | p33ING1 | p33ING1b | p47 | p47ING1a | inhibitor of growth family, member 1 | p53 interacts with ING1b. This interaction was modelled on a demonstrated interaction between p53 from an unspecified species and human ING1b. | BIND | 12208736 |
| ING4 | MGC12557 | my036 | p29ING4 | inhibitor of growth family, member 4 | - | HPRD,BioGRID | 12750254 |
| ING4 | MGC12557 | my036 | p29ING4 | inhibitor of growth family, member 4 | p53 interacts with p29ING4. | BIND | 12750254 |
| ING5 | FLJ23842 | p28ING5 | inhibitor of growth family, member 5 | - | HPRD,BioGRID | 12750254 |
| ING5 | FLJ23842 | p28ING5 | inhibitor of growth family, member 5 | p53 interacts with p28ING5. | BIND | 12750254 |
| KAT2B | CAF | P | P/CAF | PCAF | K(lysine) acetyltransferase 2B | Biochemical Activity | BioGRID | 12068014 |
| KIAA0087 | - | KIAA0087 | Two-hybrid | BioGRID | 16169070 |
| KPNB1 | IMB1 | IPOB | Impnb | MGC2155 | MGC2156 | MGC2157 | NTF97 | karyopherin (importin) beta 1 | - | HPRD,BioGRID | 11297531 |
| LAMA4 | DKFZp686D23145 | LAMA3 | LAMA4*-1 | laminin, alpha 4 | Two-hybrid | BioGRID | 16169070 |
| MAD2L1BP | CMT2 | KIAA0110 | MGC11282 | RP1-261G23.6 | MAD2L1 binding protein | Two-hybrid | BioGRID | 16169070 |
| MAGEB18 | MGC33889 | melanoma antigen family B, 18 | Two-hybrid | BioGRID | 16189514 |
| MAPK1 | ERK | ERK2 | ERT1 | MAPK2 | P42MAPK | PRKM1 | PRKM2 | p38 | p40 | p41 | p41mapk | mitogen-activated protein kinase 1 | Affinity Capture-Western | BioGRID | 10958792 |
| MAPK10 | FLJ12099 | FLJ33785 | JNK3 | JNK3A | MGC50974 | PRKM10 | p493F12 | p54bSAPK | mitogen-activated protein kinase 10 | - | HPRD,BioGRID | 9393873 |
| MAPK3 | ERK1 | HS44KDAP | HUMKER1A | MGC20180 | P44ERK1 | P44MAPK | PRKM3 | mitogen-activated protein kinase 3 | - | HPRD,BioGRID | 10958792 |
| MAPK8 | JNK | JNK1 | JNK1A2 | JNK21B1/2 | PRKM8 | SAPK1 | mitogen-activated protein kinase 8 | - | HPRD,BioGRID | 9393873|9732264 |
| MAPK9 | JNK-55 | JNK2 | JNK2A | JNK2ALPHA | JNK2B | JNK2BETA | PRKM9 | SAPK | p54a | p54aSAPK | mitogen-activated protein kinase 9 | - | HPRD,BioGRID | 9393873 |
| MDC1 | DKFZp781A0122 | KIAA0170 | MGC166888 | NFBD1 | mediator of DNA damage checkpoint 1 | - | HPRD,BioGRID | 14519663 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | TP53 (p53) interacts with the MDM2-promoter. | BIND | 15735750 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | An unspecified isoform of HDM2 interacts with p53. This interaction was modelled on a demonstrated interaction between human HDM2 and p53 from an unspecified species. | BIND | 15950904 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | Affinity Capture-Western Biochemical Activity Co-fractionation Reconstituted Complex | BioGRID | 7686617 |9271120 |9529249 |9809062 |10722742 |10766163 |10892746 |11709713 |12208736 |12426395 |12620407 |12915590 |14612427 |14671306 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | p53 interacts with Mdm2. | BIND | 15577944 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | HDM-2 interacts with p53 | BIND | 15558054 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | p53 interacts with mdm2 promoter. | BIND | 16009130 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | MDM2 (HDM2) interacts with TP53 (p53). This interaction was modeled on a demonstrated interaction between MDM2 from an unspecified species, and human TP53. | BIND | 15688025 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | Mdm2 ubiquitinates p53. | BIND | 10822382 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | Mdm2 interacts with p53. | BIND | 11046142 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | hdm2 interacts with p53. This interaction was modeled on a demonstrated interaction between hdm2 and p53 both from an unspecified species. | BIND | 9223638 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | Mdm2 interacts with p53. | BIND | 16023600 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | An unspecified isoform of MDM2 interacts with and ubiquitinates p53. This interaction was modeled on a demonstrated interaction between MDM2 from an unspecified species and human p53. | BIND | 15933712 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | Mdm2 interacts with and ubiquitinates p53. | BIND | 15280377 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | p53 interacts with Mdm2. | BIND | 11597128 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | Mdm2 interacts with and ubiquitinates p53 | BIND | 14671306 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | p53 interacts with unspecified isoform of Mdm2. | BIND | 15824742 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | p53 interacts with and is ubiquitinated by Mdm2. | BIND | 15577914 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | - | HPRD | 7686617 |8058315 |10721693 |11709713 |12372616 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | Hdm2 interacts with p53. | BIND | 15210108 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | An unspecifed isoform of Mdm2 interacts with p53. | BIND | 15867431 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | Mdm2 ubiquitinates p53. This interaction was modeled on a demonstrated interaction between mouse Mdm2 and human p53. | BIND | 15242646 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | Mdm2 interacts with p53. | BIND | 10562557 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | p53 interacts with mdm2. This interaction was modelled on a demonstrated interaction between human p53 and mdm2 from an unspecified species. | BIND | 10781812 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | p53 interacts with the hdm2 promoter. | BIND | 15525514 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | An unspecified isoform of Mdm2 ubiquitinates p53. This interaction was modeled on a demonstrated interaction between Mdm2 and p53 both from an unspecified species. | BIND | 15963787 |
| MDM4 | DKFZp781B1423 | HDMX | MDMX | MGC132766 | MRP1 | Mdm4 p53 binding protein homolog (mouse) | Biochemical Activity Reconstituted Complex | BioGRID | 9226370 |12393902 |
| MDM4 | DKFZp781B1423 | HDMX | MDMX | MGC132766 | MRP1 | Mdm4 p53 binding protein homolog (mouse) | - | HPRD | 8895579 |9226370 |11528400 |12393902 |
| MED1 | CRSP1 | CRSP200 | DRIP205 | DRIP230 | MGC71488 | PBP | PPARBP | PPARGBP | RB18A | TRAP220 | TRIP2 | mediator complex subunit 1 | - | HPRD,BioGRID | 9444950 |11118038 |
| MED17 | CRSP6 | CRSP77 | DRIP80 | FLJ10812 | TRAP80 | mediator complex subunit 17 | - | HPRD,BioGRID | 10198638 |
| MNAT1 | MAT1 | RNF66 | TFB3 | menage a trois homolog 1, cyclin H assembly factor (Xenopus laevis) | - | HPRD,BioGRID | 9372954 |
| MPHOSPH6 | MPP | MPP-6 | MPP6 | M-phase phosphoprotein 6 | Two-hybrid | BioGRID | 16169070 |
| MRE11A | ATLD | HNGS1 | MRE11 | MRE11B | MRE11 meiotic recombination 11 homolog A (S. cerevisiae) | Mre11 interacts with p53. This interaction was modeled on a demonstrated interaction between Mre11 and p53 both from African green monkey. | BIND | 15806145 |
| MSX1 | HOX7 | HYD1 | msh homeobox 1 | MSX1 interacts with TP53 (p53). This interaction was modeled on a demonstrated interaction between MSX1 from an unspecified species, and human p53. | BIND | 15705871 |
| MTA1 | - | metastasis associated 1 | - | HPRD,BioGRID | 12920132 |
| MTA2 | DKFZp686F2281 | MTA1L1 | PID | metastasis associated 1 family, member 2 | Reconstituted Complex | BioGRID | 12920132 |
| MTA2 | DKFZp686F2281 | MTA1L1 | PID | metastasis associated 1 family, member 2 | PID interacts with p53. | BIND | 11099047 |
| MUC1 | CD227 | EMA | H23AG | MAM6 | PEM | PEMT | PUM | mucin 1, cell surface associated | MUC1 C-ter interacts with p53. | BIND | 15710329 |
| NCL | C23 | FLJ45706 | nucleolin | - | HPRD,BioGRID | 12138209 |
| NDN | HsT16328 | PWCR | necdin homolog (mouse) | - | HPRD,BioGRID | 10347180 |
| NFKBIA | IKBA | MAD-3 | NFKBI | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | - | HPRD,BioGRID | 11799106 |
| NP | FLJ94043 | FLJ97288 | FLJ97312 | MGC117396 | MGC125915 | MGC125916 | PNP | PRO1837 | PUNP | nucleoside phosphorylase | Two-hybrid | BioGRID | 16169070 |
| NPM1 | B23 | MGC104254 | NPM | nucleophosmin (nucleolar phosphoprotein B23, numatrin) | - | HPRD | 12080348 |
| NR3C1 | GCCR | GCR | GR | GRL | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | - | HPRD,BioGRID | 11080152 |
| NR3C1 | GCCR | GCR | GR | GRL | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | p53 interacts with GR. This interaction was modelled on a demonstrated interaction between p53 from an unspecified species and GR from an unspecified species. | BIND | 9215863 |
| NTHL1 | NTH1 | OCTS3 | nth endonuclease III-like 1 (E. coli) | NTH1 interacts with p53. | BIND | 15358233 |
| PAFAH1B3 | - | platelet-activating factor acetylhydrolase, isoform Ib, gamma subunit 29kDa | Two-hybrid | BioGRID | 16169070 |
| PARC | DKFZp686G1042 | DKFZp686P2024 | H7AP1 | RP3-330M21.2 | p53-associated parkin-like cytoplasmic protein | - | HPRD,BioGRID | 12526791 |
| PARC | DKFZp686G1042 | DKFZp686P2024 | H7AP1 | RP3-330M21.2 | p53-associated parkin-like cytoplasmic protein | Parc interacts with p53 | BIND | 15558054 |
| PARP1 | ADPRT | ADPRT1 | PARP | PARP-1 | PPOL | pADPRT-1 | poly (ADP-ribose) polymerase 1 | - | HPRD,BioGRID | 9565608 |
| PCDHA4 | CNR1 | CNRN1 | CRNR1 | MGC138307 | MGC142169 | PCDH-ALPHA4 | protocadherin alpha 4 | Two-hybrid | BioGRID | 16169070 |
| PHB | PHB1 | prohibitin | Affinity Capture-Western Co-localization Reconstituted Complex | BioGRID | 14500729 |
| PIAS1 | DDXBP1 | GBP | GU/RH-II | MGC141878 | MGC141879 | ZMIZ3 | protein inhibitor of activated STAT, 1 | Biochemical Activity Reconstituted Complex Two-hybrid | BioGRID | 10380882 |11583632 |
| PIAS1 | DDXBP1 | GBP | GU/RH-II | MGC141878 | MGC141879 | ZMIZ3 | protein inhibitor of activated STAT, 1 | PIAS1 interacts with p53. This interaction was modeled on a demonstrated interaction between human PIAS1 and p53 from an unspecified species. | BIND | 15133049 |
| PIAS1 | DDXBP1 | GBP | GU/RH-II | MGC141878 | MGC141879 | ZMIZ3 | protein inhibitor of activated STAT, 1 | - | HPRD | 11583632 |11788578 |11867732 |15133049 |
| PIN1 | DOD | UBL5 | peptidylprolyl cis/trans isomerase, NIMA-interacting 1 | - | HPRD,BioGRID | 12388558 |12397362 |
| PLAGL1 | DKFZp781P1017 | LOT1 | MGC126275 | MGC126276 | ZAC | ZAC1 | pleiomorphic adenoma gene-like 1 | - | HPRD,BioGRID | 11360197 |
| PLK3 | CNK | FNK | PRK | polo-like kinase 3 (Drosophila) | - | HPRD,BioGRID | 11551930 |
| PML | MYL | PP8675 | RNF71 | TRIM19 | promyelocytic leukemia | Affinity Capture-Western Reconstituted Complex | BioGRID | 11025664 |11080164 |12915590 |
| PML | MYL | PP8675 | RNF71 | TRIM19 | promyelocytic leukemia | - | HPRD | 11025664 |11080164 |11704853 |
| POLA1 | DKFZp686K1672 | POLA | p180 | polymerase (DNA directed), alpha 1, catalytic subunit | - | HPRD,BioGRID | 11917009 |
| PPA1 | IOPPP | MGC111556 | PP | PP1 | SID6-8061 | pyrophosphatase (inorganic) 1 | Two-hybrid | BioGRID | 16169070 |
| PPP1R13L | IASPP | NKIP1 | RAI | protein phosphatase 1, regulatory (inhibitor) subunit 13 like | iASPP interacts with p53 | BIND | 15558054 |
| PPP1R13L | IASPP | NKIP1 | RAI | protein phosphatase 1, regulatory (inhibitor) subunit 13 like | - | HPRD,BioGRID | 12524540 |
| PPP2CA | PP2Ac | PP2CA | RP-C | protein phosphatase 2 (formerly 2A), catalytic subunit, alpha isoform | p53 interacts with PPP2CA. This interaction is modeled on a demonstrated interaction between p53 from an unspecified source and human PP2AC. | BIND | 12556559 |
| PPP4C | PP4 | PPH3 | PPX | protein phosphatase 4 (formerly X), catalytic subunit | Reconstituted Complex | BioGRID | 9837938 |
| PRKCA | AAG6 | MGC129900 | MGC129901 | PKC-alpha | PKCA | PRKACA | protein kinase C, alpha | - | HPRD | 9935181 |
| PRKDC | DNA-PKcs | DNAPK | DNPK1 | HYRC | HYRC1 | XRCC7 | p350 | protein kinase, DNA-activated, catalytic polypeptide | - | HPRD | 10470151 |
| PRKDC | DNA-PKcs | DNAPK | DNPK1 | HYRC | HYRC1 | XRCC7 | p350 | protein kinase, DNA-activated, catalytic polypeptide | Biochemical Activity Protein-peptide | BioGRID | 9679063 |10608806 |12756247 |
| PRKRA | DYT16 | HSD14 | PACT | RAX | protein kinase, interferon-inducible double stranded RNA dependent activator | - | HPRD,BioGRID | 9010216 |
| PRKRIR | DAP4 | MGC102750 | P52rIPK | protein-kinase, interferon-inducible double stranded RNA dependent inhibitor, repressor of (P58 repressor) | Reconstituted Complex | BioGRID | 12384512 |
| PRMT1 | ANM1 | HCP1 | HRMT1L2 | IR1B4 | protein arginine methyltransferase 1 | - | HPRD,BioGRID | 15186775 |
| PRMT1 | ANM1 | HCP1 | HRMT1L2 | IR1B4 | protein arginine methyltransferase 1 | PRMT1 interacts with p53. This interaction was modeled on a demonstrated interaction between rat PRMT1 and human p53. | BIND | 15186775 |
| PSMD11 | MGC3844 | Rpn6 | S9 | p44.5 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 | Two-hybrid | BioGRID | 16169070 |
| PTEN | 10q23del | BZS | MGC11227 | MHAM | MMAC1 | PTEN1 | TEP1 | phosphatase and tensin homolog | - | HPRD,BioGRID | 12620407 |
| PTGS2 | COX-2 | COX2 | GRIPGHS | PGG/HS | PGHS-2 | PHS-2 | hCox-2 | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | - | HPRD,BioGRID | 11687965 |
| PTGS2 | COX-2 | COX2 | GRIPGHS | PGG/HS | PGHS-2 | PHS-2 | hCox-2 | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | COX-2 interacts with p53. | BIND | 15608668 |
| PTTG1 | EAP1 | HPTTG | MGC126883 | MGC138276 | PTTG | TUTR1 | pituitary tumor-transforming 1 | - | HPRD,BioGRID | 12355087 |
| PTTG1 | EAP1 | HPTTG | MGC126883 | MGC138276 | PTTG | TUTR1 | pituitary tumor-transforming 1 | p53 interacts with securin. | BIND | 12355087 |
| RAB4A | HRES-1/RAB4 | RAB4 | RAB4A, member RAS oncogene family | Two-hybrid | BioGRID | 16169070 |
| RAD51 | BRCC5 | HRAD51 | HsRad51 | HsT16930 | RAD51A | RECA | RAD51 homolog (RecA homolog, E. coli) (S. cerevisiae) | - | HPRD,BioGRID | 8617246 |9380510 |
| RCHY1 | ARNIP | CHIMP | DKFZp586C1620 | PIRH2 | PRO1996 | RNF199 | ZNF363 | hARNIP | ring finger and CHY zinc finger domain containing 1 | - | HPRD,BioGRID | 12654245 |
| RECQL4 | RECQ4 | RecQ protein-like 4 | TP53 (p53) interacts with the RECQL4 (RECQ4) promoter. | BIND | 15674334 |
| RFC1 | A1 | MGC51786 | MHCBFB | PO-GA | RECC1 | RFC | RFC140 | replication factor C (activator 1) 1, 145kDa | Affinity Capture-Western | BioGRID | 12509469 |
| RFWD2 | COP1 | FLJ10416 | RNF200 | ring finger and WD repeat domain 2 | The ubiquitin ligase COP1 interacts with p53 to cause p53 degradation. | BIND | 15103385 |
| RPA1 | HSSB | REPA1 | RF-A | RP-A | RPA70 | replication protein A1, 70kDa | Affinity Capture-Western Reconstituted Complex | BioGRID | 11751427 |15489903 |
| RPL11 | GIG34 | ribosomal protein L11 | Affinity Capture-Western | BioGRID | 14612427 |
| RRM2 | R2 | RR2M | ribonucleotide reductase M2 polypeptide | - | HPRD,BioGRID | 12615712 |
| RRM2B | DKFZp686M05248 | MGC102856 | MGC42116 | p53R2 | ribonucleotide reductase M2 B (TP53 inducible) | - | HPRD,BioGRID | 12615712 |
| S100A4 | 18A2 | 42A | CAPL | FSP1 | MTS1 | P9KA | PEL98 | S100 calcium binding protein A4 | - | HPRD,BioGRID | 11278647 |11527429 |12942774 |15116098 |
| S100A8 | 60B8AG | CAGA | CFAG | CGLA | CP-10 | L1Ag | MA387 | MIF | MRP8 | NIF | P8 | S100 calcium binding protein A8 | Two-hybrid | BioGRID | 16169070 |
| S100B | NEF | S100 | S100beta | S100 calcium binding protein B | Affinity Capture-Western | BioGRID | 15178678 |
| S100B | NEF | S100 | S100beta | S100 calcium binding protein B | - | HPRD | 10876243 |11527429 |
| SAE1 | AOS1 | FLJ3091 | HSPC140 | SUA1 | SUMO1 activating enzyme subunit 1 | - | HPRD | 15327968 |
| SAT1 | DC21 | KFSD | SAT | SSAT | SSAT-1 | spermidine/spermine N1-acetyltransferase 1 | Two-hybrid | BioGRID | 16169070 |
| SERPINB9 | CAP-3 | CAP3 | PI9 | serpin peptidase inhibitor, clade B (ovalbumin), member 9 | Two-hybrid | BioGRID | 16169070 |
| SETD7 | FLJ21193 | KIAA1717 | KMT7 | SET7 | SET7/9 | SET9 | SET domain containing (lysine methyltransferase) 7 | - | HPRD | 15525938 |
| SFN | YWHAS | stratifin | Affinity Capture-Western | BioGRID | 11896572 |
| SFN | YWHAS | stratifin | p53 interacts with the SFN (14-3-3-sigma) promoter. | BIND | 15574328 |
| SFN | YWHAS | stratifin | p53 interacts with 14-3-3-sigma promoter. | BIND | 16009130 |
| SHISA5 | SCOTIN | hShisa5 | shisa homolog 5 (Xenopus laevis) | - | HPRD | 12135983 |
| SIN3A | DKFZp434K2235 | FLJ90319 | KIAA0700 | SIN3 homolog A, transcription regulator (yeast) | p53 interacts with mSin3A. This interaction was modeled on a demonstrated interaction between human p53 and mSin3A from an unspecified species. | BIND | 10823891 |
| SMARCA4 | BAF190 | BRG1 | FLJ39786 | SNF2 | SNF2-BETA | SNF2L4 | SNF2LB | SWI2 | hSNF2b | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 | Affinity Capture-Western Co-fractionation Reconstituted Complex | BioGRID | 11950834 |
| SMARCB1 | BAF47 | INI1 | RDT | SNF5 | SNF5L1 | Sfh1p | Snr1 | hSNFS | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1 | - | HPRD,BioGRID | 11950834 |
| SMARCC1 | BAF155 | CRACC1 | Rsc8 | SRG3 | SWI3 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 | Co-fractionation | BioGRID | 11950834 |
| SMG1 | 61E3.4 | ATX | KIAA0421 | LIP | PI-3-kinase-related kinase SMG-1 | hSMG-1 phosphorylates p53. | BIND | 15175154 |
| SMN1 | BCD541 | SMA | SMA1 | SMA2 | SMA3 | SMA4 | SMA@ | SMN | SMNT | T-BCD541 | survival of motor neuron 1, telomeric | - | HPRD,BioGRID | 11704667 |
| SNRPN | DKFZp686C0927 | DKFZp686M12165 | DKFZp761I1912 | DKFZp762N022 | FLJ33569 | FLJ36996 | FLJ39265 | HCERN3 | MGC29886 | PWCR | RT-LI | SM-D | SMN | SNRNP-N | SNURF-SNRPN | small nuclear ribonucleoprotein polypeptide N | Two-hybrid | BioGRID | 16169070 |
| SP1 | - | Sp1 transcription factor | p53 interacts with Sp1. | BIND | 15574328 |
| SP1 | - | Sp1 transcription factor | p53 interacts with Sp1. | BIND | 9492043 |
| SP1 | - | Sp1 transcription factor | TP53 (p53) interacts with Sp1. | BIND | 9685344 |
| SP1 | - | Sp1 transcription factor | TP53 (p53) interacts with SP1. | BIND | 15674334 |
| SSTR3 | - | somatostatin receptor 3 | - | HPRD | 8961277 |
| STK11 | LKB1 | PJS | serine/threonine kinase 11 | - | HPRD,BioGRID | 11430832 |
| STK4 | DKFZp686A2068 | KRS2 | MST1 | YSK3 | serine/threonine kinase 4 | - | HPRD | 12384512 |12942774 |
| STX5 | SED5 | STX5A | syntaxin 5 | Two-hybrid | BioGRID | 16169070 |
| SULT1E1 | EST | EST-1 | MGC34459 | STE | sulfotransferase family 1E, estrogen-preferring, member 1 | Two-hybrid | BioGRID | 16169070 |
| SUMO1 | DAP-1 | GMP1 | OFC10 | PIC1 | SENP2 | SMT3 | SMT3C | SMT3H3 | SUMO-1 | UBL1 | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | Two-hybrid | BioGRID | 10961991 |
| SUMO1 | DAP-1 | GMP1 | OFC10 | PIC1 | SENP2 | SMT3 | SMT3C | SMT3H3 | SUMO-1 | UBL1 | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | p53 interacts with SUMO-1. This interaction was modelled on a demonstrated interaction between p53 from human and SUMO-1 from an unspecified species. | BIND | 10788439 |
| TADA3L | ADA3 | FLJ20221 | FLJ21329 | hADA3 | transcriptional adaptor 3 (NGG1 homolog, yeast)-like | - | HPRD,BioGRID | 11707411 |
| TAF1 | BA2R | CCG1 | CCGS | DYT3 | KAT4 | N-TAF1 | NSCL2 | OF | P250 | TAF2A | TAFII250 | TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa | - | HPRD | 7809597 |
| TAF1 | BA2R | CCG1 | CCGS | DYT3 | KAT4 | N-TAF1 | NSCL2 | OF | P250 | TAF2A | TAFII250 | TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa | Affinity Capture-Western | BioGRID | 10359315 |
| TAF1A | MGC:17061 | RAFI48 | SL1 | TAFI48 | TATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa | - | HPRD,BioGRID | 10913176 |
| TAF1B | MGC:9349 | RAF1B | RAFI63 | SL1 | TAFI63 | TATA box binding protein (TBP)-associated factor, RNA polymerase I, B, 63kDa | - | HPRD,BioGRID | 10913176 |
| TAF1C | MGC:39976 | SL1 | TAFI110 | TAFI95 | TATA box binding protein (TBP)-associated factor, RNA polymerase I, C, 110kDa | - | HPRD,BioGRID | 10913176 |
| TAF9 | AD-004 | AK6 | CGI-137 | CINAP | CIP | MGC1603 | MGC3647 | MGC5067 | MGC:1603 | MGC:3647 | MGC:5067 | TAF2G | TAFII31 | TAFII32 | TAFIID32 | TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa | - | HPRD,BioGRID | 7761466 |
| TAF9 | AD-004 | AK6 | CGI-137 | CINAP | CIP | MGC1603 | MGC3647 | MGC5067 | MGC:1603 | MGC:3647 | MGC:5067 | TAF2G | TAFII31 | TAFII32 | TAFIID32 | TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa | - | HPRD | 7761466 |7809597 |11278372 |
| TAG | - | tumor antigen gene | Affinity Capture-Western | BioGRID | 14612521 |
| TBP | GTF2D | GTF2D1 | MGC117320 | MGC126054 | MGC126055 | SCA17 | TFIID | TATA box binding protein | - | HPRD,BioGRID | 1465435 |
| TDG | - | thymine-DNA glycosylase | Two-hybrid | BioGRID | 10961991 |
| TEP1 | TLP1 | TP1 | TROVE1 | VAULT2 | p240 | telomerase-associated protein 1 | - | HPRD,BioGRID | 10597287 |
| TFAP2A | AP-2 | AP-2alpha | AP2TF | BOFS | TFAP2 | transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) | p53 interacts with AP2alpha. | BIND | 12226108 |
| TFAP2A | AP-2 | AP-2alpha | AP2TF | BOFS | TFAP2 | transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) | - | HPRD,BioGRID | 12226108 |
| TFAP2C | AP2-GAMMA | ERF1 | TFAP2G | hAP-2g | transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) | p53 interacts with AP2gamma. This interaction was modelled on a demonstrated interaction between p53 from human and AP2gamma from an unspecified species. | BIND | 12226108 |
| TFAP2C | AP2-GAMMA | ERF1 | TFAP2G | hAP-2g | transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) | Reconstituted Complex | BioGRID | 12226108 |
| TFDP1 | DP1 | DRTF1 | Dp-1 | transcription factor Dp-1 | - | HPRD,BioGRID | 8816502 |
| THAP8 | FLJ32891 | THAP domain containing 8 | Two-hybrid | BioGRID | 16169070 |
| THRB | ERBA-BETA | ERBA2 | GRTH | MGC126109 | MGC126110 | NR1A2 | PRTH | THR1 | THRB1 | THRB2 | thyroid hormone receptor, beta (erythroblastic leukemia viral (v-erb-a) oncogene homolog 2, avian) | - | HPRD,BioGRID | 11258898 |
| TK1 | TK2 | thymidine kinase 1, soluble | Two-hybrid | BioGRID | 16169070 |
| TNFRSF10B | CD262 | DR5 | KILLER | KILLER/DR5 | TRAIL-R2 | TRAILR2 | TRICK2 | TRICK2A | TRICK2B | TRICKB | ZTNFR9 | tumor necrosis factor receptor superfamily, member 10b | p53 interacts with DR5. | BIND | 15607964 |
| TNFRSF10C | CD263 | DCR1 | LIT | MGC149501 | MGC149502 | TRAILR3 | TRID | tumor necrosis factor receptor superfamily, member 10c, decoy without an intracellular domain | TP53 (p53) interacts with TNFRSF10C (TRAIL-R3) gene. | BIND | 14623878 |
| TOP1 | TOPI | topoisomerase (DNA) I | - | HPRD,BioGRID | 10468612 |
| TOP2A | TOP2 | TP2A | topoisomerase (DNA) II alpha 170kDa | - | HPRD,BioGRID | 10666337 |
| TOP2B | TOPIIB | top2beta | topoisomerase (DNA) II beta 180kDa | - | HPRD,BioGRID | 10666337 |
| TOPORS | LUN | P53BP3 | RP31 | TP53BPL | topoisomerase I binding, arginine/serine-rich | Topors interacts with p53. This interaction was modelled on a demonstrated interaction between human Topors and mouse p53. | BIND | 11842245 |
| TOPORS | LUN | P53BP3 | RP31 | TP53BPL | topoisomerase I binding, arginine/serine-rich | Two-hybrid | BioGRID | 11842245 |
| TOPORS | LUN | P53BP3 | RP31 | TP53BPL | topoisomerase I binding, arginine/serine-rich | - | HPRD | 10415337 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | p53 interacts with itself to form a dimer, trimer and tetramer of p53. | BIND | 15674341 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | p53 exists as a dimer in solution. | BIND | 15321712 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | Co-purification Reconstituted Complex | BioGRID | 14580323 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | p53 interacts with itself. | BIND | 16009130 |
| TP53BP1 | 53BP1 | FLJ41424 | MGC138366 | p202 | tumor protein p53 binding protein 1 | Affinity Capture-Western Co-crystal Structure Co-localization Reconstituted Complex Two-hybrid | BioGRID | 8016121 |11877378 |12110597 |12351827 |14978302 |15364958 |15611139 |
| TP53BP1 | 53BP1 | FLJ41424 | MGC138366 | p202 | tumor protein p53 binding protein 1 | - | HPRD | 8016121 |12110597 |
| TP53BP2 | 53BP2 | ASPP2 | BBP | PPP1R13A | p53BP2 | tumor protein p53 binding protein, 2 | - | HPRD,BioGRID | 8016121 |8668206 |8875926 |
| TP53I3 | PIG3 | tumor protein p53 inducible protein 3 | p53 interacts with PIG3 promoter. | BIND | 16009130 |
| TP53INP1 | DKFZp434M1317 | FLJ22139 | SIP | TP53DINP1 | TP53INP1A | TP53INP1B | Teap | p53DINP1 | tumor protein p53 inducible nuclear protein 1 | - | HPRD,BioGRID | 11511362 |12851404 |
| TP53RK | BUD32 | C20orf64 | Nori-2 | Nori-2p | PRPK | dJ101A2 | TP53 regulating kinase | Reconstituted Complex | BioGRID | 12659830 |
| TSG101 | TSG10 | VPS23 | tumor susceptibility gene 101 | - | HPRD,BioGRID | 11172000 |
| TXN | DKFZp686B1993 | MGC61975 | TRX | TRX1 | thioredoxin | p53 interacts with Trx. | BIND | 15824742 |
| UBE2A | HHR6A | RAD6A | UBC2 | ubiquitin-conjugating enzyme E2A (RAD6 homolog) | - | HPRD,BioGRID | 12640129 |
| UBE2I | C358B7.1 | P18 | UBC9 | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | - | HPRD,BioGRID | 8921390 |
| UBE2I | C358B7.1 | P18 | UBC9 | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | p53 interacts with Ubc9. This interaction was modelled on a demonstrated interaction between p53 from human and Ubc9 from an unspecified species. | BIND | 10788439 |
| UBE2I | C358B7.1 | P18 | UBC9 | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | UBE2I interacts with TP53. This interaction was modelled on a demonstrated interaction between D. melanogaster Lwr (DmUbc9) and p53 from an unspecified species. | BIND | 9514881 |
| UBE2K | DKFZp564C1216 | DKFZp686J24237 | E2-25K | HIP2 | HYPG | LIG | ubiquitin-conjugating enzyme E2K (UBC1 homolog, yeast) | - | HPRD | 10634809 |
| UBE3A | ANCR | AS | E6-AP | EPVE6AP | FLJ26981 | HPVE6A | ubiquitin protein ligase E3A | - | HPRD | 9143503 |9369221 |
| USP7 | HAUSP | TEF1 | ubiquitin specific peptidase 7 (herpes virus-associated) | - | HPRD,BioGRID | 11923872 |
| USP7 | HAUSP | TEF1 | ubiquitin specific peptidase 7 (herpes virus-associated) | p53 interacts with HAUSP. | BIND | 15916963 |
| WDR33 | FLJ11294 | WDC146 | WD repeat domain 33 | Two-hybrid | BioGRID | 16169070 |
| WRN | DKFZp686C2056 | RECQ3 | RECQL2 | RECQL3 | Werner syndrome | WRN interacts with p53. | BIND | 12080066 |
| WRN | DKFZp686C2056 | RECQ3 | RECQL2 | RECQL3 | Werner syndrome | - | HPRD,BioGRID | 11427532 |12080066 |
| WT1 | GUD | WAGR | WIT-2 | WT33 | Wilms tumor 1 | - | HPRD,BioGRID | 8389468 |
| WWOX | D16S432E | FOR | FRA16D | HHCMA56 | PRO0128 | WOX1 | WW domain containing oxidoreductase | - | HPRD,BioGRID | 11058590 |
| XRCC1 | RCC | X-ray repair complementing defective repair in Chinese hamster cells 1 | Affinity Capture-Western | BioGRID | 15044383 |
| XRCC6 | CTC75 | CTCBF | G22P1 | KU70 | ML8 | TLAA | X-ray repair complementing defective repair in Chinese hamster cells 6 | TP53 (p53) interacts with XRCC6 (Ku70). | BIND | 15782130 |
| YBX1 | BP-8 | CSDA2 | CSDB | DBPB | MDR-NF1 | MGC104858 | MGC110976 | MGC117250 | NSEP-1 | NSEP1 | YB-1 | YB1 | Y box binding protein 1 | - | HPRD | 11175333 |11973333 |
| YBX1 | BP-8 | CSDA2 | CSDB | DBPB | MDR-NF1 | MGC104858 | MGC110976 | MGC117250 | NSEP-1 | NSEP1 | YB-1 | YB1 | Y box binding protein 1 | in vitro in vivo Reconstituted Complex | BioGRID | 11175333 |15136035 |
| YWHAZ | KCIP-1 | MGC111427 | MGC126532 | MGC138156 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | - | HPRD,BioGRID | 9620776 |
| YY1 | DELTA | INO80S | NF-E1 | UCRBP | YIN-YANG-1 | YY1 transcription factor | YY1 interacts with p53. | BIND | 15210108 |
| ZCCHC10 | FLJ20094 | zinc finger, CCHC domain containing 10 | Two-hybrid | BioGRID | 16169070 |
| ZHX1 | - | zinc fingers and homeoboxes 1 | - | HPRD | 15383276 |
| ZNF148 | BERF-1 | BFCOL1 | HT-BETA | ZBP-89 | ZFP148 | pHZ-52 | zinc finger protein 148 | - | HPRD,BioGRID | 11416144 |
| ZNF24 | KOX17 | RSG-A | ZNF191 | ZSCAN3 | Zfp191 | zinc finger protein 24 | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG CELL CYCLE | 128 | 84 | All SZGR 2.0 genes in this pathway |
| KEGG P53 SIGNALING PATHWAY | 69 | 45 | All SZGR 2.0 genes in this pathway |
| KEGG APOPTOSIS | 88 | 62 | All SZGR 2.0 genes in this pathway |
| KEGG WNT SIGNALING PATHWAY | 151 | 112 | All SZGR 2.0 genes in this pathway |
| KEGG NEUROTROPHIN SIGNALING PATHWAY | 126 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG AMYOTROPHIC LATERAL SCLEROSIS ALS | 53 | 43 | All SZGR 2.0 genes in this pathway |
| KEGG HUNTINGTONS DISEASE | 185 | 109 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG COLORECTAL CANCER | 62 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG PANCREATIC CANCER | 70 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG ENDOMETRIAL CANCER | 52 | 45 | All SZGR 2.0 genes in this pathway |
| KEGG GLIOMA | 65 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG PROSTATE CANCER | 89 | 75 | All SZGR 2.0 genes in this pathway |
| KEGG THYROID CANCER | 29 | 26 | All SZGR 2.0 genes in this pathway |
| KEGG BASAL CELL CARCINOMA | 55 | 44 | All SZGR 2.0 genes in this pathway |
| KEGG MELANOMA | 71 | 57 | All SZGR 2.0 genes in this pathway |
| KEGG BLADDER CANCER | 42 | 33 | All SZGR 2.0 genes in this pathway |
| KEGG CHRONIC MYELOID LEUKEMIA | 73 | 59 | All SZGR 2.0 genes in this pathway |
| KEGG SMALL CELL LUNG CANCER | 84 | 67 | All SZGR 2.0 genes in this pathway |
| KEGG NON SMALL CELL LUNG CANCER | 54 | 47 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CHEMICAL PATHWAY | 22 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ATM PATHWAY | 20 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA G1 PATHWAY | 28 | 21 | All SZGR 2.0 genes in this pathway |
| BIOCARTA G2 PATHWAY | 24 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TID PATHWAY | 19 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CTCF PATHWAY | 23 | 18 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RNA PATHWAY | 10 | 9 | All SZGR 2.0 genes in this pathway |
| BIOCARTA P53HYPOXIA PATHWAY | 23 | 21 | All SZGR 2.0 genes in this pathway |
| BIOCARTA P53 PATHWAY | 16 | 13 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RB PATHWAY | 13 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PML PATHWAY | 17 | 12 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ATRBRCA PATHWAY | 21 | 12 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TEL PATHWAY | 18 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ARF PATHWAY | 17 | 13 | All SZGR 2.0 genes in this pathway |
| SA G1 AND S PHASES | 15 | 10 | All SZGR 2.0 genes in this pathway |
| ST JNK MAPK PATHWAY | 40 | 30 | All SZGR 2.0 genes in this pathway |
| ST FAS SIGNALING PATHWAY | 65 | 54 | All SZGR 2.0 genes in this pathway |
| PID HDAC CLASSIII PATHWAY | 25 | 20 | All SZGR 2.0 genes in this pathway |
| PID MYC ACTIV PATHWAY | 79 | 62 | All SZGR 2.0 genes in this pathway |
| PID LKB1 PATHWAY | 47 | 37 | All SZGR 2.0 genes in this pathway |
| PID REG GR PATHWAY | 82 | 60 | All SZGR 2.0 genes in this pathway |
| PID P53 DOWNSTREAM PATHWAY | 137 | 94 | All SZGR 2.0 genes in this pathway |
| PID P75 NTR PATHWAY | 69 | 51 | All SZGR 2.0 genes in this pathway |
| PID AP1 PATHWAY | 70 | 60 | All SZGR 2.0 genes in this pathway |
| PID HIF1A PATHWAY | 19 | 12 | All SZGR 2.0 genes in this pathway |
| PID P38 ALPHA BETA DOWNSTREAM PATHWAY | 38 | 29 | All SZGR 2.0 genes in this pathway |
| PID AURORA A PATHWAY | 31 | 20 | All SZGR 2.0 genes in this pathway |
| PID BARD1 PATHWAY | 29 | 19 | All SZGR 2.0 genes in this pathway |
| PID P53 REGULATION PATHWAY | 59 | 50 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | 29 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME PRE NOTCH EXPRESSION AND PROCESSING | 44 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE CHECKPOINTS | 124 | 70 | All SZGR 2.0 genes in this pathway |
| REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | 57 | 35 | All SZGR 2.0 genes in this pathway |
| REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | 51 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | 132 | 101 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH | 103 | 64 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOSIS | 148 | 94 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF BH3 ONLY PROTEINS | 17 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | 30 | 21 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| LIU PROSTATE CANCER UP | 96 | 57 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 48HR UP | 128 | 95 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 96HR UP | 117 | 84 | All SZGR 2.0 genes in this pathway |
| LAU APOPTOSIS CDKN2A UP | 55 | 40 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS A UP | 191 | 128 | All SZGR 2.0 genes in this pathway |
| CAIRO PML TARGETS BOUND BY MYC UP | 23 | 17 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| MARKS HDAC TARGETS UP | 23 | 15 | All SZGR 2.0 genes in this pathway |
| MARKS ACETYLATED NON HISTONE PROTEINS | 15 | 9 | All SZGR 2.0 genes in this pathway |
| WAKASUGI HAVE ZNF143 BINDING SITES | 58 | 33 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 XPCS UP | 28 | 16 | All SZGR 2.0 genes in this pathway |
| TURJANSKI MAPK1 AND MAPK2 TARGETS | 12 | 10 | All SZGR 2.0 genes in this pathway |
| TURJANSKI MAPK8 AND MAPK9 TARGETS | 8 | 7 | All SZGR 2.0 genes in this pathway |
| TURJANSKI MAPK14 TARGETS | 10 | 10 | All SZGR 2.0 genes in this pathway |
| GALLUZZI PERMEABILIZE MITOCHONDRIA | 43 | 31 | All SZGR 2.0 genes in this pathway |
| DUTTA APOPTOSIS VIA NFKB | 33 | 25 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE AUGMENTED BY MYC | 108 | 74 | All SZGR 2.0 genes in this pathway |
| HONRADO BREAST CANCER BRCA1 VS BRCA2 | 18 | 12 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 UP | 309 | 199 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS NEUROEPITHELIUM UP | 176 | 111 | All SZGR 2.0 genes in this pathway |
| JAZAG TGFB1 SIGNALING VIA SMAD4 UP | 108 | 66 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| PUJANA BREAST CANCER LIT INT NETWORK | 101 | 73 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| KHETCHOUMIAN TRIM24 TARGETS UP | 47 | 38 | All SZGR 2.0 genes in this pathway |
| SCIAN CELL CYCLE TARGETS OF TP53 AND TP73 UP | 9 | 7 | All SZGR 2.0 genes in this pathway |
| SCIAN INVERSED TARGETS OF TP53 AND TP73 UP | 11 | 9 | All SZGR 2.0 genes in this pathway |
| FRIDMAN SENESCENCE UP | 77 | 60 | All SZGR 2.0 genes in this pathway |
| FRIDMAN IMMORTALIZATION DN | 34 | 24 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION DN | 206 | 136 | All SZGR 2.0 genes in this pathway |
| HE PTEN TARGETS UP | 16 | 15 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| DING LUNG CANCER MUTATED SIGNIFICANTLY | 26 | 22 | All SZGR 2.0 genes in this pathway |
| DING LUNG CANCER MUTATED FREQUENTLY | 12 | 10 | All SZGR 2.0 genes in this pathway |
| SHEPARD CRUSH AND BURN MUTANT UP | 197 | 110 | All SZGR 2.0 genes in this pathway |
| CHEOK RESPONSE TO MERCAPTOPURINE UP | 13 | 10 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO UP | 205 | 126 | All SZGR 2.0 genes in this pathway |
| REN BOUND BY E2F | 61 | 40 | All SZGR 2.0 genes in this pathway |
| PEART HDAC PROLIFERATION CLUSTER DN | 76 | 57 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE UP | 203 | 130 | All SZGR 2.0 genes in this pathway |
| HASLINGER B CLL WITH 17P13 DELETION | 21 | 15 | All SZGR 2.0 genes in this pathway |
| PARK HSC AND MULTIPOTENT PROGENITORS | 50 | 33 | All SZGR 2.0 genes in this pathway |
| NGUYEN NOTCH1 TARGETS DN | 86 | 67 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS BY TBH AND H2O2 | 36 | 24 | All SZGR 2.0 genes in this pathway |
| NATSUME RESPONSE TO INTERFERON BETA UP | 71 | 49 | All SZGR 2.0 genes in this pathway |
| ONGUSAHA TP53 TARGETS | 38 | 23 | All SZGR 2.0 genes in this pathway |
| MARIADASON RESPONSE TO CURCUMIN SULINDAC 7 | 19 | 10 | All SZGR 2.0 genes in this pathway |
| GEISS RESPONSE TO DSRNA DN | 16 | 8 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| WU HBX TARGETS 3 DN | 13 | 9 | All SZGR 2.0 genes in this pathway |
| MOREIRA RESPONSE TO TSA UP | 28 | 23 | All SZGR 2.0 genes in this pathway |
| MODY HIPPOCAMPUS PRENATAL | 42 | 33 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| SANSOM APC MYC TARGETS | 217 | 138 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS REQUIRE MYC | 210 | 123 | All SZGR 2.0 genes in this pathway |
| SANSOM WNT PATHWAY REQUIRE MYC | 58 | 43 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY OVERCONNECTED IN BREAST CANCER | 22 | 19 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE DN | 315 | 197 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| CADWELL ATG16L1 TARGETS DN | 70 | 43 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB UP | 245 | 159 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA METAGENE | 242 | 168 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| JIANG TIP30 TARGETS DN | 24 | 14 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| MOOTHA MITOCHONDRIA | 447 | 277 | All SZGR 2.0 genes in this pathway |
| DING LUNG CANCER MUTATED RECURRENTLY | 6 | 5 | All SZGR 2.0 genes in this pathway |
| DING LUNG CANCER BY MUTATION RATE | 20 | 18 | All SZGR 2.0 genes in this pathway |
| TCGA GLIOBLASTOMA MUTATED | 8 | 8 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA UP | 207 | 143 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC UP | 72 | 53 | All SZGR 2.0 genes in this pathway |
| DANG MYC TARGETS UP | 143 | 100 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| WONG EMBRYONIC STEM CELL CORE | 335 | 193 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 16 | 79 | 47 | All SZGR 2.0 genes in this pathway |
| NIELSEN MALIGNAT FIBROUS HISTIOCYTOMA DN | 18 | 11 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO GONADOTROPHINS DN | 87 | 66 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO FORSKOLIN DN | 88 | 68 | All SZGR 2.0 genes in this pathway |
| STAMBOLSKY TARGETS OF MUTATED TP53 UP | 49 | 35 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 UP | 140 | 94 | All SZGR 2.0 genes in this pathway |
| DUAN PRDM5 TARGETS | 79 | 52 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 7 | 403 | 240 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR BOUND ES | 462 | 273 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 | 227 | 149 | All SZGR 2.0 genes in this pathway |