Gene Page: NRIP1
Summary ?
| GeneID | 8204 |
| Symbol | NRIP1 |
| Synonyms | RIP140 |
| Description | nuclear receptor interacting protein 1 |
| Reference | MIM:602490|HGNC:HGNC:8001|Ensembl:ENSG00000180530|HPRD:03927|Vega:OTTHUMG00000074323 |
| Gene type | protein-coding |
| Map location | 21q11.2 |
| Pascal p-value | 4.662E-4 |
| Sherlock p-value | 0.53 |
| Fetal beta | -0.064 |
| eGene | Putamen basal ganglia |
| Support | CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Girard_2011 | Whole Exome Sequencing analysis | The data set included 15 DNMs found from the exomes of 14 schizophrenia probands and their parents. | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NRIP1 | chr21 | 16338349 | T | G | NM_003489 | p.722K>T | missense | Schizophrenia | DNM:Girard_2011 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2822746 | 21 | 15899364 | NRIP1 | ENSG00000180530.5 | 1.06941E-6 | 0.01 | 537957 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
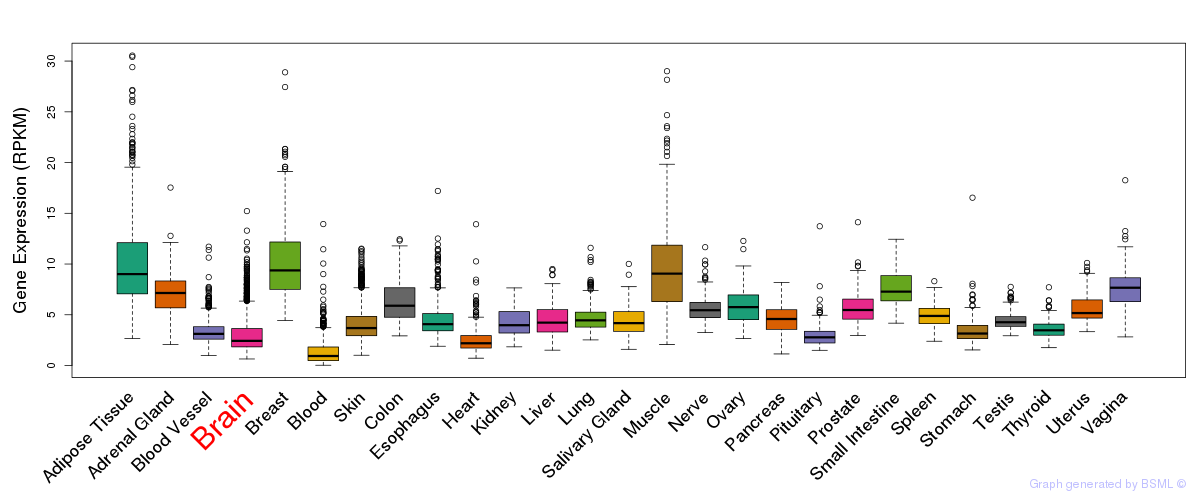
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005102 | receptor binding | IEA | Neurotransmitter (GO term level: 4) | - |
| GO:0003713 | transcription coactivator activity | IDA | 7641693 |10364267 |11518808 | |
| GO:0003713 | transcription coactivator activity | NAS | 15572661 | |
| GO:0003714 | transcription corepressor activity | IDA | 10364267 | |
| GO:0042826 | histone deacetylase binding | IEA | - | |
| GO:0035257 | nuclear hormone receptor binding | IPI | 11518808 | |
| GO:0030331 | estrogen receptor binding | IPI | 7641693 | |
| GO:0035259 | glucocorticoid receptor binding | IPI | 10364267 |11266503 |12773562 | |
| GO:0050681 | androgen receptor binding | NAS | 15572661 | |
| GO:0046965 | retinoid X receptor binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | IDA | 10364267 | |
| GO:0001543 | ovarian follicle rupture | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0019915 | sequestering of lipid | IEA | - | |
| GO:0030728 | ovulation | IEA | - | |
| GO:0030521 | androgen receptor signaling pathway | NAS | 15572661 | |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | IDA | 7641693 |10364267 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000118 | histone deacetylase complex | IEA | - | |
| GO:0005634 | nucleus | IDA | 7641693 |11266503 |12773562 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AHR | - | aryl hydrocarbon receptor | - | HPRD,BioGRID | 10428779 |
| CCDC85B | DIPA | coiled-coil domain containing 85B | Two-hybrid | BioGRID | 16189514 |
| CEP70 | BITE | FLJ13036 | centrosomal protein 70kDa | Two-hybrid | BioGRID | 16189514 |
| CTBP1 | BARS | MGC104684 | C-terminal binding protein 1 | Reconstituted Complex | BioGRID | 15060175 |
| CTBP1 | BARS | MGC104684 | C-terminal binding protein 1 | - | HPRD | 11509661 |
| CTBP2 | - | C-terminal binding protein 2 | Reconstituted Complex Two-hybrid | BioGRID | 15060175 |16189514 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | ESR1 (ER-alpha) interacts with NRIP1 (RIP140). This interaction was modelled on a demonstrated interaction between human ESR1 and mouse NRIP1. | BIND | 9774463 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | ER interacts with NRIP-1 enhancer. | BIND | 16009131 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | - | HPRD,BioGRID | 7641693 |
| FOXA1 | HNF3A | MGC33105 | TCF3A | forkhead box A1 | FOXA1 interacts with NRIP-1 enhancer. | BIND | 16009131 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | - | HPRD | 11006275|11285237 |
| HDAC5 | FLJ90614 | HD5 | NY-CO-9 | histone deacetylase 5 | Co-localization Reconstituted Complex | BioGRID | 15060175 |
| HDAC9 | DKFZp779K1053 | HD7 | HDAC | HDAC7 | HDAC7B | HDAC9B | HDAC9FL | HDRP | KIAA0744 | MITR | histone deacetylase 9 | Reconstituted Complex | BioGRID | 15060175 |
| JUN | AP-1 | AP1 | c-Jun | jun oncogene | - | HPRD,BioGRID | 12554755 |
| KAT2B | CAF | P | P/CAF | PCAF | K(lysine) acetyltransferase 2B | - | HPRD | 14581481 |
| LDOC1 | BCUR1 | Mar7 | Mart7 | leucine zipper, down-regulated in cancer 1 | Two-hybrid | BioGRID | 16189514 |
| MAP3K7 | TAK1 | TGF1a | mitogen-activated protein kinase kinase kinase 7 | - | HPRD | 9556573 |
| NR0B1 | AHC | AHCH | AHX | DAX-1 | DAX1 | DSS | GTD | HHG | NROB1 | nuclear receptor subfamily 0, group B, member 1 | Reconstituted Complex Two-hybrid | BioGRID | 11459805 |
| NR1H3 | LXR-a | LXRA | RLD-1 | nuclear receptor subfamily 1, group H, member 3 | - | HPRD | 10022764 |
| NR2C1 | TR2 | nuclear receptor subfamily 2, group C, member 1 | - | HPRD | 9774688 |
| NR3C1 | GCCR | GCR | GR | GRL | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | Affinity Capture-Western Reconstituted Complex | BioGRID | 10364267 |11266503 |12773562 |
| NR3C1 | GCCR | GCR | GR | GRL | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | - | HPRD | 10364267 |12773562 |
| NR5A1 | AD4BP | ELP | FTZ1 | FTZF1 | SF-1 | SF1 | nuclear receptor subfamily 5, group A, member 1 | - | HPRD,BioGRID | 11459805 |12782406 |
| POLR2A | MGC75453 | POLR2 | POLRA | RPB1 | RPBh1 | RPO2 | RPOL2 | RpIILS | hRPB220 | hsRPB1 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | Pol II interacts with NRIP-1 enhancer. | BIND | 16009131 |
| PPARA | MGC2237 | MGC2452 | NR1C1 | PPAR | hPPAR | peroxisome proliferator-activated receptor alpha | - | HPRD,BioGRID | 9626662 |10022764 |
| PPARG | NR1C3 | PPARG1 | PPARG2 | PPARgamma | peroxisome proliferator-activated receptor gamma | - | HPRD,BioGRID | 9626662 |
| RARA | NR1B1 | RAR | retinoic acid receptor, alpha | Affinity Capture-Western Reconstituted Complex | BioGRID | 8887632 |12549917 |14581481 |
| RARA | NR1B1 | RAR | retinoic acid receptor, alpha | - | HPRD | 12403842 |12549917 |
| RXRA | FLJ00280 | FLJ00318 | FLJ16020 | FLJ16733 | MGC102720 | NR2B1 | retinoid X receptor, alpha | - | HPRD,BioGRID | 8887632 |
| RXRB | DAUDI6 | H-2RIIBP | MGC1831 | NR2B2 | RCoR-1 | retinoid X receptor, beta | - | HPRD | 12403842 |12549917 |
| RXRB | DAUDI6 | H-2RIIBP | MGC1831 | NR2B2 | RCoR-1 | retinoid X receptor, beta | Reconstituted Complex | BioGRID | 9626662 |
| TEX11 | TGC1 | TSGA3 | testis expressed 11 | Two-hybrid | BioGRID | 16189514 |
| THRA | AR7 | EAR7 | ERB-T-1 | ERBA | ERBA1 | MGC000261 | MGC43240 | NR1A1 | THRA1 | THRA2 | c-ERBA-1 | thyroid hormone receptor, alpha (erythroblastic leukemia viral (v-erb-a) oncogene homolog, avian) | Reconstituted Complex | BioGRID | 9626662 |
| TNFRSF14 | ATAR | HVEA | HVEM | LIGHTR | TR2 | tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) | - | HPRD | 9774688 |
| TRAF2 | MGC:45012 | TRAP | TRAP3 | TNF receptor-associated factor 2 | Two-hybrid | BioGRID | 16189514 |
| YWHAH | YWHA1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide | - | HPRD | 11266503 |
| YWHAQ | 14-3-3 | 1C5 | HS1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide | Co-localization Phenotypic Suppression Reconstituted Complex | BioGRID | 11266503 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA CARM ER PATHWAY | 35 | 27 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PPARA PATHWAY | 58 | 43 | All SZGR 2.0 genes in this pathway |
| PID AR PATHWAY | 61 | 46 | All SZGR 2.0 genes in this pathway |
| PID ERA GENOMIC PATHWAY | 65 | 37 | All SZGR 2.0 genes in this pathway |
| PID RETINOIC ACID PATHWAY | 30 | 23 | All SZGR 2.0 genes in this pathway |
| PID HNF3A PATHWAY | 44 | 29 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN | 463 | 290 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL DN | 60 | 39 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS UP | 372 | 227 | All SZGR 2.0 genes in this pathway |
| WANG CLIM2 TARGETS UP | 269 | 146 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION ERYTHROCYTE UP | 157 | 104 | All SZGR 2.0 genes in this pathway |
| LEE NEURAL CREST STEM CELL UP | 146 | 99 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL UP | 316 | 190 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML DIVIDING VS NORMAL QUIESCENT DN | 95 | 57 | All SZGR 2.0 genes in this pathway |
| GRAHAM NORMAL QUIESCENT VS NORMAL DIVIDING UP | 66 | 47 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 3 4WK DN | 39 | 27 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 5 6WK DN | 137 | 97 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 6 7WK UP | 197 | 135 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL TGFB1 TARGETS UP | 169 | 127 | All SZGR 2.0 genes in this pathway |
| MCBRYAN TERMINAL END BUD DN | 7 | 6 | All SZGR 2.0 genes in this pathway |
| MARTIN INTERACT WITH HDAC | 44 | 31 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR UP | 487 | 286 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT AND CANCER BOX4 DN | 32 | 22 | All SZGR 2.0 genes in this pathway |
| FRASOR RESPONSE TO ESTRADIOL UP | 37 | 28 | All SZGR 2.0 genes in this pathway |
| BECKER TAMOXIFEN RESISTANCE UP | 50 | 36 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| HASLINGER B CLL WITH MUTATED VH GENES | 18 | 14 | All SZGR 2.0 genes in this pathway |
| WANG IMMORTALIZED BY HOXA9 AND MEIS1 DN | 24 | 15 | All SZGR 2.0 genes in this pathway |
| STOSSI RESPONSE TO ESTRADIOL | 50 | 35 | All SZGR 2.0 genes in this pathway |
| FERRANDO T ALL WITH MLL ENL FUSION UP | 87 | 67 | All SZGR 2.0 genes in this pathway |
| BROWN MYELOID CELL DEVELOPMENT DN | 129 | 86 | All SZGR 2.0 genes in this pathway |
| ROETH TERT TARGETS UP | 14 | 11 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA MS UP | 48 | 32 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION UP | 314 | 201 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| RUAN RESPONSE TO TNF TROGLITAZONE DN | 42 | 24 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C2 | 54 | 39 | All SZGR 2.0 genes in this pathway |
| RUAN RESPONSE TO TNF DN | 84 | 50 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 5 | 126 | 78 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D2 | 41 | 30 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| GEORGANTAS HSC MARKERS | 71 | 47 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK DN | 318 | 220 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS UP | 280 | 183 | All SZGR 2.0 genes in this pathway |
| DURCHDEWALD SKIN CARCINOGENESIS DN | 264 | 168 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 4 | 307 | 185 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER T4 | 94 | 69 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| ZHENG FOXP3 TARGETS IN T LYMPHOCYTE DN | 37 | 29 | All SZGR 2.0 genes in this pathway |
| STEARMAN LUNG CANCER EARLY VS LATE UP | 125 | 89 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE DN | 258 | 160 | All SZGR 2.0 genes in this pathway |
| MASSARWEH RESPONSE TO ESTRADIOL | 61 | 47 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BRAIN DN | 85 | 58 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| IZADPANAH STEM CELL ADIPOSE VS BONE DN | 108 | 68 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| MATZUK OVULATION | 14 | 10 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C CLUSTER DN | 32 | 21 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C DN | 59 | 39 | All SZGR 2.0 genes in this pathway |
| FERRARI RESPONSE TO FENRETINIDE UP | 22 | 16 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 12 | 30 | 20 | All SZGR 2.0 genes in this pathway |
| MARSON FOXP3 TARGETS UP | 66 | 43 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA PCA3 UP | 80 | 54 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| RUAN RESPONSE TO TROGLITAZONE DN | 19 | 14 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| HOELZEL NF1 TARGETS UP | 139 | 93 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 6HR UP | 229 | 149 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR UP | 324 | 193 | All SZGR 2.0 genes in this pathway |
| CERIBELLI GENES INACTIVE AND BOUND BY NFY | 45 | 27 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS DN | 229 | 135 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| IKEDA MIR30 TARGETS UP | 116 | 87 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-103/107 | 2969 | 2975 | 1A | hsa-miR-103brain | AGCAGCAUUGUACAGGGCUAUGA |
| hsa-miR-107brain | AGCAGCAUUGUACAGGGCUAUCA | ||||
| miR-125/351 | 2238 | 2244 | 1A | hsa-miR-125bbrain | UCCCUGAGACCCUAACUUGUGA |
| hsa-miR-125abrain | UCCCUGAGACCCUUUAACCUGUG | ||||
| miR-128 | 244 | 250 | 1A | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-143 | 1308 | 1314 | 1A | hsa-miR-143brain | UGAGAUGAAGCACUGUAGCUCA |
| miR-186 | 622 | 628 | 1A | hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU |
| miR-192/215 | 2234 | 2240 | m8 | hsa-miR-192 | CUGACCUAUGAAUUGACAGCC |
| hsa-miR-215 | AUGACCUAUGAAUUGACAGAC | ||||
| miR-193 | 2039 | 2045 | m8 | hsa-miR-193a | AACUGGCCUACAAAGUCCCAG |
| hsa-miR-193b | AACUGGCCCUCAAAGUCCCGCUUU | ||||
| miR-194 | 2726 | 2732 | 1A | hsa-miR-194 | UGUAACAGCAACUCCAUGUGGA |
| miR-200bc/429 | 3060 | 3066 | m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-208 | 3022 | 3028 | 1A | hsa-miR-208 | AUAAGACGAGCAAAAAGCUUGU |
| miR-214 | 2967 | 2973 | m8 | hsa-miR-214brain | ACAGCAGGCACAGACAGGCAG |
| miR-219 | 2076 | 2082 | m8 | hsa-miR-219brain | UGAUUGUCCAAACGCAAUUCU |
| miR-25/32/92/363/367 | 1201 | 1207 | 1A | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| miR-26 | 1842 | 1849 | 1A,m8 | hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC |
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
| miR-27 | 244 | 250 | m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-30-5p | 3292 | 3299 | 1A,m8 | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG | ||||
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-324-3p | 3445 | 3451 | 1A | hsa-miR-324-3p | CCACUGCCCCAGGUGCUGCUGG |
| miR-328 | 1385 | 1391 | m8 | hsa-miR-328brain | CUGGCCCUCUCUGCCCUUCCGU |
| miR-33 | 2812 | 2818 | 1A | hsa-miR-33 | GUGCAUUGUAGUUGCAUUG |
| hsa-miR-33b | GUGCAUUGCUGUUGCAUUGCA | ||||
| miR-377 | 3397 | 3403 | 1A | hsa-miR-377 | AUCACACAAAGGCAACUUUUGU |
| miR-378 | 1312 | 1318 | 1A | hsa-miR-378 | CUCCUGACUCCAGGUCCUGUGU |
| miR-380-5p | 3151 | 3158 | 1A,m8 | hsa-miR-380-5p | UGGUUGACCAUAGAACAUGCGC |
| hsa-miR-563 | AGGUUGACAUACGUUUCCC | ||||
| miR-448 | 149 | 155 | m8 | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| miR-488 | 1296 | 1302 | m8 | hsa-miR-488 | CCCAGAUAAUGGCACUCUCAA |
| miR-490 | 1769 | 1776 | 1A,m8 | hsa-miR-490 | CAACCUGGAGGACUCCAUGCUG |
| miR-496 | 2763 | 2769 | 1A | hsa-miR-496 | AUUACAUGGCCAAUCUC |
| miR-499 | 3022 | 3028 | 1A | hsa-miR-499 | UUAAGACUUGCAGUGAUGUUUAA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.