Gene Page: RGS5
Summary ?
| GeneID | 8490 |
| Symbol | RGS5 |
| Synonyms | MST092|MST106|MST129|MSTP032|MSTP092|MSTP106|MSTP129 |
| Description | regulator of G-protein signaling 5 |
| Reference | MIM:603276|HGNC:HGNC:10001|Ensembl:ENSG00000143248|Ensembl:ENSG00000232995|HPRD:04471|Vega:OTTHUMG00000034441 |
| Gene type | protein-coding |
| Map location | 1q23.1 |
| Sherlock p-value | 0.511 |
| Fetal beta | -1.873 |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00814 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0036 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2498500 | chr1 | 119673092 | RGS5 | 8490 | 0.06 | trans | ||
| rs4027083 | chr1 | 234452908 | RGS5 | 8490 | 0.03 | trans | ||
| rs4665370 | chr2 | 21794755 | RGS5 | 8490 | 0.14 | trans | ||
| rs693742 | chr3 | 127494235 | RGS5 | 8490 | 0.14 | trans | ||
| rs6440537 | chr3 | 148079187 | RGS5 | 8490 | 0.08 | trans | ||
| rs4129734 | chr3 | 148107826 | RGS5 | 8490 | 0.13 | trans | ||
| snp_a-2214158 | 0 | RGS5 | 8490 | 0.1 | trans | |||
| rs7814325 | chr8 | 2655993 | RGS5 | 8490 | 0.18 | trans | ||
| rs17057381 | chr8 | 27416800 | RGS5 | 8490 | 0.17 | trans | ||
| rs12263767 | chr10 | 67526911 | RGS5 | 8490 | 0 | trans | ||
| rs12251350 | chr10 | 67529955 | RGS5 | 8490 | 0.03 | trans | ||
| rs17127155 | chr10 | 111943519 | RGS5 | 8490 | 0.02 | trans | ||
| snp_a-1895744 | 0 | RGS5 | 8490 | 0.1 | trans | |||
| rs2000658 | chr11 | 125590392 | RGS5 | 8490 | 0.16 | trans | ||
| rs411816 | chr12 | 27627804 | RGS5 | 8490 | 0.13 | trans | ||
| rs11175122 | chr12 | 64084446 | RGS5 | 8490 | 0.01 | trans | ||
| rs9526072 | chr13 | 32883582 | RGS5 | 8490 | 0 | trans | ||
| rs2760312 | chr13 | 101106737 | RGS5 | 8490 | 0.1 | trans | ||
| rs6498459 | chr16 | 13795898 | RGS5 | 8490 | 0.14 | trans | ||
| rs6530359 | chrX | 9957034 | RGS5 | 8490 | 0.16 | trans |
Section II. Transcriptome annotation
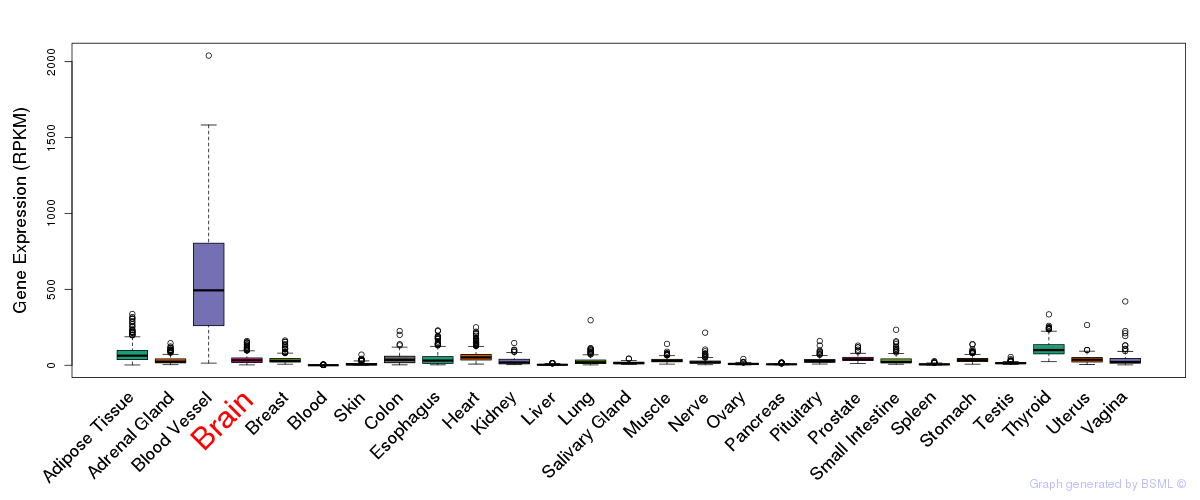
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HSDL2 | 0.83 | 0.77 |
| SEPT2 | 0.81 | 0.77 |
| PLOD2 | 0.80 | 0.78 |
| MSI2 | 0.80 | 0.76 |
| PPAP2B | 0.79 | 0.81 |
| SNX5 | 0.78 | 0.75 |
| PDLIM5 | 0.77 | 0.80 |
| SUCLG2 | 0.76 | 0.77 |
| FUT10 | 0.76 | 0.78 |
| PSAT1 | 0.76 | 0.79 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C3orf54 | -0.35 | -0.35 |
| AC011491.1 | -0.35 | -0.41 |
| RTN4RL1 | -0.35 | -0.27 |
| LRFN2 | -0.33 | -0.27 |
| MPPED1 | -0.33 | -0.24 |
| CABYR | -0.33 | -0.28 |
| RASGEF1C | -0.32 | -0.23 |
| RTN2 | -0.32 | -0.35 |
| DYNC1I1 | -0.31 | -0.26 |
| RPRM | -0.31 | -0.29 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004871 | signal transducer activity | IEA | - | |
| GO:0005096 | GTPase activator activity | TAS | 10471929 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0009968 | negative regulation of signal transduction | IEA | - | |
| GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway | TAS | 10471929 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | IDA | 18029348 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| WINTER HYPOXIA DN | 52 | 30 | All SZGR 2.0 genes in this pathway |
| PICCALUGA ANGIOIMMUNOBLASTIC LYMPHOMA UP | 205 | 140 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| BERTUCCI MEDULLARY VS DUCTAL BREAST CANCER DN | 169 | 118 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION DN | 329 | 219 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES SKIN UP | 177 | 113 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 5 6WK DN | 137 | 97 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2B | 392 | 251 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER BASAL VS LULMINAL | 330 | 215 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM4 | 261 | 153 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| BROWN MYELOID CELL DEVELOPMENT DN | 129 | 86 | All SZGR 2.0 genes in this pathway |
| KANG IMMORTALIZED BY TERT DN | 102 | 67 | All SZGR 2.0 genes in this pathway |
| RORIE TARGETS OF EWSR1 FLI1 FUSION DN | 30 | 23 | All SZGR 2.0 genes in this pathway |
| WESTON VEGFA TARGETS 6HR | 59 | 38 | All SZGR 2.0 genes in this pathway |
| WESTON VEGFA TARGETS | 108 | 71 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN UP | 181 | 112 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| CADWELL ATG16L1 TARGETS DN | 70 | 43 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| CHEN LIVER METABOLISM QTL CIS | 93 | 40 | All SZGR 2.0 genes in this pathway |
| KUROKAWA LIVER CANCER EARLY RECURRENCE UP | 12 | 9 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 4 | 15 | 11 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO GONADOTROPHINS DN | 87 | 66 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO FORSKOLIN DN | 88 | 68 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| PANGAS TUMOR SUPPRESSION BY SMAD1 AND SMAD5 UP | 134 | 85 | All SZGR 2.0 genes in this pathway |
| KATSANOU ELAVL1 TARGETS UP | 169 | 105 | All SZGR 2.0 genes in this pathway |
| PLASARI NFIC TARGETS BASAL UP | 27 | 17 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 SIGNALING VIA NFIC 1HR DN | 106 | 77 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 SIGNALING VIA NFIC 10HR UP | 54 | 38 | All SZGR 2.0 genes in this pathway |
| GUILLAUMOND KLF10 TARGETS UP | 51 | 39 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-186 | 2608 | 2614 | m8 | hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.