Gene Page: RUNX1
Summary ?
| GeneID | 861 |
| Symbol | RUNX1 |
| Synonyms | AML1|AML1-EVI-1|AMLCR1|CBF2alpha|CBFA2|EVI-1|PEBP2aB|PEBP2alpha |
| Description | runt related transcription factor 1 |
| Reference | MIM:151385|HGNC:HGNC:10471|Ensembl:ENSG00000159216|HPRD:01043|Vega:OTTHUMG00000086299 |
| Gene type | protein-coding |
| Map location | 21q22.3 |
| Pascal p-value | 0.395 |
| Fetal beta | -0.111 |
| DMG | 2 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg11498607 | 21 | 36399226 | RUNX1 | 5.647E-4 | -0.456 | 0.049 | DMG:Wockner_2014 |
| cg22866426 | 21 | 36263347 | RUNX1 | 2E-9 | -0.013 | 1.64E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2300384 | chr21 | 35253332 | RUNX1 | 861 | 0.04 | cis | ||
| rs12564871 | chr1 | 163771926 | RUNX1 | 861 | 0.2 | trans | ||
| rs6430976 | chr2 | 128822384 | RUNX1 | 861 | 0 | trans | ||
| rs11684252 | chr2 | 128829365 | RUNX1 | 861 | 0.11 | trans | ||
| rs17036588 | chr4 | 158561487 | RUNX1 | 861 | 5.435E-4 | trans | ||
| rs7445750 | chr5 | 104331223 | RUNX1 | 861 | 0.02 | trans | ||
| rs544338 | chr5 | 104625129 | RUNX1 | 861 | 0.18 | trans | ||
| rs1031612 | chr8 | 4728687 | RUNX1 | 861 | 0.12 | trans | ||
| rs509390 | chr8 | 102500402 | RUNX1 | 861 | 0.07 | trans | ||
| rs17618655 | chr9 | 73983104 | RUNX1 | 861 | 1.896E-5 | trans | ||
| rs11255704 | chr10 | 8501820 | RUNX1 | 861 | 0.07 | trans | ||
| rs4934985 | chr10 | 33905260 | RUNX1 | 861 | 0.1 | trans | ||
| rs16933426 | chr10 | 78113005 | RUNX1 | 861 | 0.01 | trans | ||
| rs17111364 | chr10 | 97901813 | RUNX1 | 861 | 0.17 | trans | ||
| rs11214244 | chr11 | 112434429 | RUNX1 | 861 | 0.15 | trans | ||
| rs11216730 | chr11 | 117918701 | RUNX1 | 861 | 0.12 | trans | ||
| rs17089379 | chr13 | 64433308 | RUNX1 | 861 | 0.05 | trans | ||
| rs8072151 | chr17 | 50228268 | RUNX1 | 861 | 4.62E-4 | trans | ||
| rs8107683 | chr19 | 35901140 | RUNX1 | 861 | 0.09 | trans | ||
| rs2179733 | chr20 | 53876875 | RUNX1 | 861 | 0.01 | trans | ||
| rs1344430 | chr20 | 60099390 | RUNX1 | 861 | 0.05 | trans | ||
| rs6652005 | 0 | RUNX1 | 861 | 0.06 | trans | |||
| rs2984344 | chrX | 71523613 | RUNX1 | 861 | 0.12 | trans | ||
| rs17340169 | chrX | 138724917 | RUNX1 | 861 | 0.13 | trans |
Section II. Transcriptome annotation
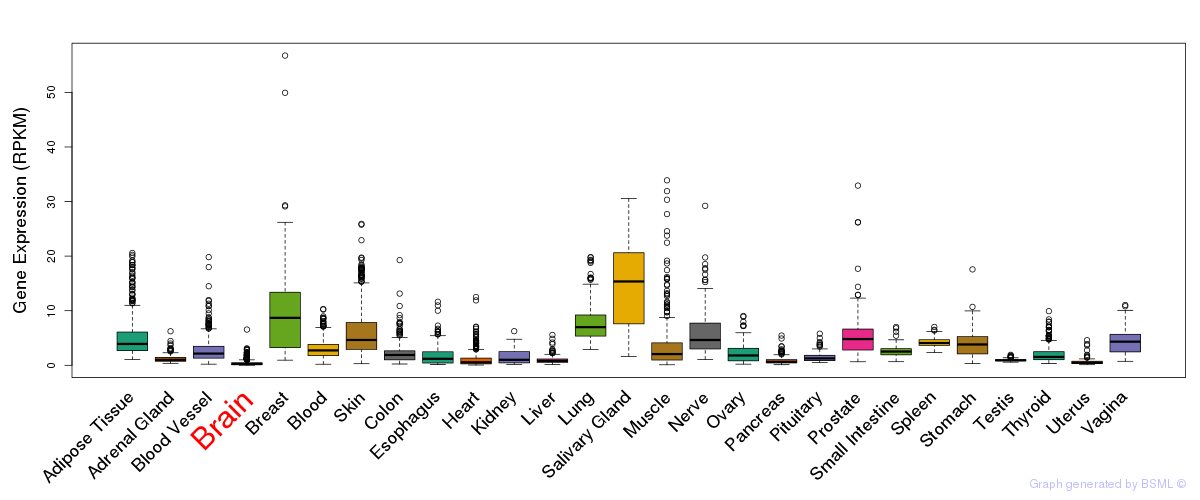
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PDCL | 0.90 | 0.90 |
| WDR3 | 0.90 | 0.90 |
| MAPRE1 | 0.90 | 0.89 |
| AP3M1 | 0.89 | 0.91 |
| RCBTB2 | 0.89 | 0.90 |
| ZFP161 | 0.89 | 0.89 |
| SUPT16H | 0.89 | 0.89 |
| SMAD4 | 0.89 | 0.91 |
| HNRNPU | 0.89 | 0.90 |
| ZBTB10 | 0.88 | 0.91 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FXYD1 | -0.67 | -0.81 |
| MT-CO2 | -0.66 | -0.82 |
| AF347015.31 | -0.66 | -0.80 |
| AIFM3 | -0.65 | -0.71 |
| C5orf53 | -0.65 | -0.69 |
| IFI27 | -0.65 | -0.79 |
| HLA-F | -0.65 | -0.70 |
| AF347015.27 | -0.65 | -0.78 |
| AF347015.33 | -0.64 | -0.77 |
| S100B | -0.63 | -0.73 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AES | AES-1 | AES-2 | ESP1 | GRG | GRG5 | TLE5 | amino-terminal enhancer of split | Two-hybrid | BioGRID | 10825294 |
| CBFA2T2 | DKFZp313F2116 | EHT | MTGR1 | ZMYND3 | core-binding factor, runt domain, alpha subunit 2; translocated to, 2 | - | HPRD | 10675041 |
| CBFA2T3 | ETO2 | MTG16 | MTGR2 | ZMYND4 | core-binding factor, runt domain, alpha subunit 2; translocated to, 3 | - | HPRD | 14703694 |
| CBFB | PEBP2B | core-binding factor, beta subunit | - | HPRD,BioGRID | 10856244 |
| CBFB | PEBP2B | core-binding factor, beta subunit | Cbf-beta interacts with AML1a. This interaction was modeled on a demonstrated interaction between mouse Cbf-beta and human AML1a. | BIND | 9751710 |
| CBFB | PEBP2B | core-binding factor, beta subunit | Cbf-beta interacts with AML1b. This interaction was modeled on a demonstrated interaction between mouse Cbf-beta and human AML1b. | BIND | 9751710 |
| CEBPB | C/EBP-beta | CRP2 | IL6DBP | LAP | MGC32080 | NF-IL6 | TCF5 | CCAAT/enhancer binding protein (C/EBP), beta | - | HPRD,BioGRID | 8622667 |
| ELF2 | EU32 | NERF | NERF-1A | NERF-1B | NERF-1a,b | NERF-2 | E74-like factor 2 (ets domain transcription factor) | - | HPRD | 14970218 |
| ELF4 | ELFR | MEF | E74-like factor 4 (ets domain transcription factor) | - | HPRD,BioGRID | 10207087 |
| FOS | AP-1 | C-FOS | v-fos FBJ murine osteosarcoma viral oncogene homolog | - | HPRD,BioGRID | 11641401 |
| JUN | AP-1 | AP1 | c-Jun | jun oncogene | - | HPRD,BioGRID | 11274169 |
| LEF1 | DKFZp586H0919 | TCF1ALPHA | lymphoid enhancer-binding factor 1 | Reconstituted Complex | BioGRID | 12551949 |
| MYST4 | DKFZp313G1618 | FLJ90335 | KAT6B | KIAA0383 | MORF | MOZ2 | qkf | querkopf | MYST histone acetyltransferase (monocytic leukemia) 4 | - | HPRD,BioGRID | 11965546 |
| PAX5 | BSAP | paired box 5 | - | HPRD,BioGRID | 10455134 |
| SMAD1 | BSP1 | JV4-1 | JV41 | MADH1 | MADR1 | SMAD family member 1 | Affinity Capture-Western | BioGRID | 10531362 |
| SMAD2 | JV18 | JV18-1 | MADH2 | MADR2 | MGC22139 | MGC34440 | hMAD-2 | hSMAD2 | SMAD family member 2 | Affinity Capture-Western | BioGRID | 10531362 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | Affinity Capture-Western | BioGRID | 10531362 |
| SMAD5 | DKFZp781C1895 | DKFZp781O1323 | Dwfc | JV5-1 | MADH5 | SMAD family member 5 | Affinity Capture-Western | BioGRID | 10531362 |
| SUV39H1 | KMT1A | MG44 | SUV39H | suppressor of variegation 3-9 homolog 1 (Drosophila) | - | HPRD,BioGRID | 12917624 |
| TLE1 | ESG | ESG1 | GRG1 | transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila) | AML1a interacts with TLE1. | BIND | 9751710 |
| TLE1 | ESG | ESG1 | GRG1 | transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila) | - | HPRD,BioGRID | 9751710 |
| TLE1 | ESG | ESG1 | GRG1 | transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila) | AML1b interacts with TLE1. | BIND | 9751710 |
| TLE2 | ESG | ESG2 | FLJ41188 | GRG2 | transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) | - | HPRD | 10825294 |
| VDR | NR1I1 | vitamin D (1,25- dihydroxyvitamin D3) receptor | Affinity Capture-Western Reconstituted Complex | BioGRID | 12460926 |
| YAP1 | YAP | YAP2 | YAP65 | YKI | Yes-associated protein 1, 65kDa | - | HPRD,BioGRID | 10228168 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG CHRONIC MYELOID LEUKEMIA | 73 | 59 | All SZGR 2.0 genes in this pathway |
| KEGG ACUTE MYELOID LEUKEMIA | 60 | 47 | All SZGR 2.0 genes in this pathway |
| PID SMAD2 3NUCLEAR PATHWAY | 82 | 63 | All SZGR 2.0 genes in this pathway |
| PID DELTA NP63 PATHWAY | 47 | 34 | All SZGR 2.0 genes in this pathway |
| BERTUCCI MEDULLARY VS DUCTAL BREAST CANCER DN | 169 | 118 | All SZGR 2.0 genes in this pathway |
| SOTIRIOU BREAST CANCER GRADE 1 VS 3 DN | 52 | 34 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP | 579 | 346 | All SZGR 2.0 genes in this pathway |
| WIKMAN ASBESTOS LUNG CANCER DN | 28 | 13 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY UP | 430 | 232 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL UP | 233 | 161 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 DN | 242 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 DN | 281 | 186 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC UP | 185 | 126 | All SZGR 2.0 genes in this pathway |
| ODONNELL TFRC TARGETS UP | 456 | 228 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 3 4WK UP | 214 | 144 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK UP | 271 | 175 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 5 6WK DN | 137 | 97 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 6 7WK UP | 197 | 135 | All SZGR 2.0 genes in this pathway |
| VETTER TARGETS OF PRKCA AND ETS1 UP | 16 | 8 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL TGFB1 TARGETS UP | 169 | 127 | All SZGR 2.0 genes in this pathway |
| KLEIN TARGETS OF BCR ABL1 FUSION | 45 | 34 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| KIM MYCL1 AMPLIFICATION TARGETS DN | 20 | 12 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM5 | 94 | 59 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| ROPERO HDAC2 TARGETS | 114 | 71 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS DN | 357 | 212 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL CIS | 128 | 77 | All SZGR 2.0 genes in this pathway |
| FERRANDO TAL1 NEIGHBORS | 21 | 15 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| OKUMURA INFLAMMATORY RESPONSE LPS | 183 | 115 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE UP | 203 | 130 | All SZGR 2.0 genes in this pathway |
| FAELT B CLL WITH VH REARRANGEMENTS UP | 48 | 34 | All SZGR 2.0 genes in this pathway |
| NUMATA CSF3 SIGNALING VIA STAT3 | 22 | 17 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED UP | 183 | 111 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 16HR DN | 86 | 62 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 10HR DN | 56 | 37 | All SZGR 2.0 genes in this pathway |
| TRAYNOR RETT SYNDROM DN | 19 | 14 | All SZGR 2.0 genes in this pathway |
| RAMASWAMY METASTASIS DN | 61 | 47 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC DN | 228 | 146 | All SZGR 2.0 genes in this pathway |
| DELASERNA MYOD TARGETS UP | 89 | 51 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 8 HR DN | 53 | 40 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 24 HR DN | 91 | 64 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D2 | 41 | 30 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| HAN JNK SINGALING UP | 35 | 21 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL DN | 128 | 93 | All SZGR 2.0 genes in this pathway |
| JAZAERI BREAST CANCER BRCA1 VS BRCA2 UP | 49 | 28 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 12HR DN | 101 | 64 | All SZGR 2.0 genes in this pathway |
| BILD MYC ONCOGENIC SIGNATURE | 206 | 117 | All SZGR 2.0 genes in this pathway |
| BILD HRAS ONCOGENIC SIGNATURE | 261 | 166 | All SZGR 2.0 genes in this pathway |
| DURCHDEWALD SKIN CARCINOGENESIS DN | 264 | 168 | All SZGR 2.0 genes in this pathway |
| SARRIO EPITHELIAL MESENCHYMAL TRANSITION UP | 180 | 114 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS UP | 170 | 107 | All SZGR 2.0 genes in this pathway |
| MCCABE BOUND BY HOXC6 | 469 | 239 | All SZGR 2.0 genes in this pathway |
| MCCABE HOXC6 TARGETS CANCER UP | 31 | 23 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K27ME3 DN | 228 | 114 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN UP | 439 | 257 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL A UP | 84 | 52 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL OPTIMAL DEBULKING | 246 | 152 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP | 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 DN | 245 | 150 | All SZGR 2.0 genes in this pathway |
| HINATA NFKB TARGETS KERATINOCYTE DN | 23 | 15 | All SZGR 2.0 genes in this pathway |
| HOFMANN MYELODYSPLASTIC SYNDROM LOW RISK UP | 22 | 15 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 15 | 31 | 19 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 4HR DN | 254 | 158 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G1 S | 147 | 76 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| KASLER HDAC7 TARGETS 1 UP | 194 | 133 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| DUAN PRDM5 TARGETS | 79 | 52 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION UP | 271 | 165 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR UP | 199 | 143 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 3 TRANSIENTLY INDUCED BY EGF | 222 | 159 | All SZGR 2.0 genes in this pathway |