Gene Page: TIMELESS
Summary ?
| GeneID | 8914 |
| Symbol | TIMELESS |
| Synonyms | TIM|TIM1|hTIM |
| Description | timeless circadian clock |
| Reference | MIM:603887|HGNC:HGNC:11813|Ensembl:ENSG00000111602|HPRD:04864|Vega:OTTHUMG00000170600 |
| Gene type | protein-coding |
| Map location | 12q13.3 |
| Pascal p-value | 0.002 |
| Fetal beta | 1.034 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 1 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| ch.12.1183517F | 12 | 56819834 | TIMELESS | 1.97E-5 | -0.646 | 0.016 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs4757560 | chr11 | 17690161 | TIMELESS | 8914 | 0.07 | trans |
Section II. Transcriptome annotation
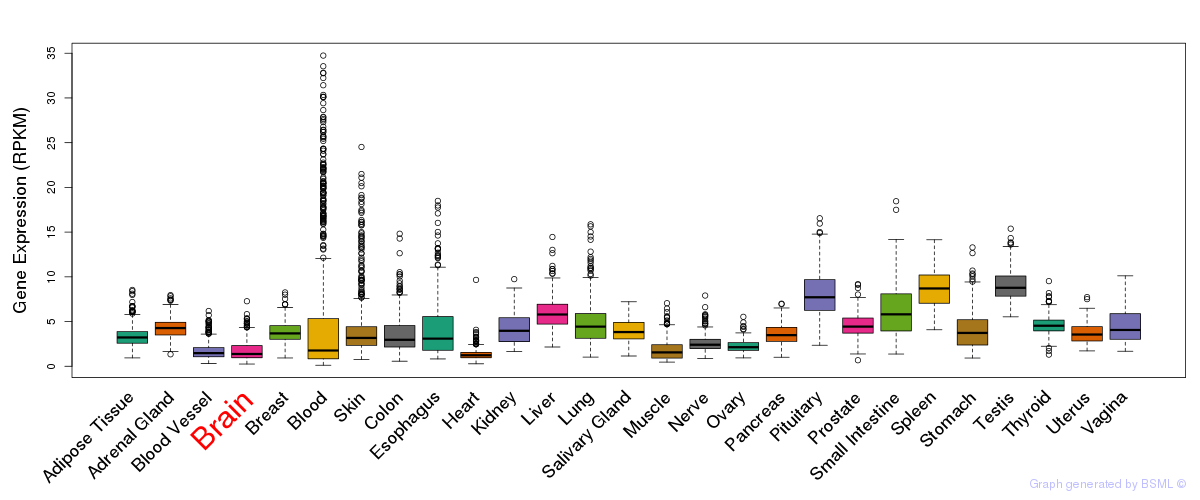
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PON2 | 0.55 | 0.53 |
| C10orf54 | 0.54 | 0.59 |
| ACOX2 | 0.53 | 0.57 |
| B2M | 0.53 | 0.57 |
| SRGN | 0.52 | 0.61 |
| GIMAP2 | 0.52 | 0.52 |
| TMBIM1 | 0.52 | 0.60 |
| NMI | 0.52 | 0.50 |
| CXCL16 | 0.52 | 0.56 |
| BBOX1 | 0.52 | 0.54 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RTF1 | -0.46 | -0.49 |
| DPF1 | -0.46 | -0.51 |
| BOP1 | -0.46 | -0.50 |
| CHD3 | -0.46 | -0.51 |
| GTF2F1 | -0.46 | -0.49 |
| SAFB | -0.46 | -0.48 |
| AC011498.3 | -0.45 | -0.49 |
| ZNF643 | -0.45 | -0.49 |
| NOVA2 | -0.45 | -0.49 |
| NCL | -0.45 | -0.49 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | ISS | - | |
| GO:0046982 | protein heterodimerization activity | ISS | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0002009 | morphogenesis of an epithelium | ISS | - | |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0007049 | cell cycle | IEA | - | |
| GO:0007067 | mitosis | IEA | - | |
| GO:0009582 | detection of abiotic stimulus | TAS | 9856466 | |
| GO:0007623 | circadian rhythm | ISS | - | |
| GO:0006974 | response to DNA damage stimulus | IEA | - | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0016481 | negative regulation of transcription | IDA | 9856465 | |
| GO:0033261 | regulation of S phase | IMP | 17102137 | |
| GO:0042127 | regulation of cell proliferation | IMP | 17102137 | |
| GO:0051301 | cell division | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000790 | nuclear chromatin | IDA | 17102137 | |
| GO:0005634 | nucleus | IC | 9856465 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID ATR PATHWAY | 39 | 25 | All SZGR 2.0 genes in this pathway |
| PID CIRCADIAN PATHWAY | 16 | 15 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 UP | 201 | 125 | All SZGR 2.0 genes in this pathway |
| SOTIRIOU BREAST CANCER GRADE 1 VS 3 UP | 151 | 84 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| DITTMER PTHLH TARGETS DN | 73 | 51 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| NADERI BREAST CANCER PROGNOSIS UP | 50 | 26 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| JAZAG TGFB1 SIGNALING UP | 108 | 69 | All SZGR 2.0 genes in this pathway |
| PUJANA XPRSS INT NETWORK | 168 | 103 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA CENTERED NETWORK | 117 | 72 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 1 DN | 163 | 115 | All SZGR 2.0 genes in this pathway |
| HASLINGER B CLL WITH CHROMOSOME 12 TRISOMY | 24 | 12 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS REQUIRE MYC | 210 | 123 | All SZGR 2.0 genes in this pathway |
| FUJII YBX1 TARGETS DN | 202 | 132 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| GRADE COLON AND RECTAL CANCER UP | 285 | 167 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| FRASOR RESPONSE TO SERM OR FULVESTRANT DN | 50 | 29 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR DN | 185 | 116 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR DN | 277 | 166 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS DN | 89 | 50 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| KOBAYASHI EGFR SIGNALING 24HR DN | 251 | 151 | All SZGR 2.0 genes in this pathway |
| FOURNIER ACINAR DEVELOPMENT LATE 2 | 277 | 172 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| WINNEPENNINCKX MELANOMA METASTASIS UP | 162 | 86 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS GROWING | 243 | 155 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR UP | 324 | 193 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
| KATSANOU ELAVL1 TARGETS DN | 148 | 88 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| ZHOU CELL CYCLE GENES IN IR RESPONSE 6HR | 85 | 49 | All SZGR 2.0 genes in this pathway |
| ZHOU CELL CYCLE GENES IN IR RESPONSE 24HR | 128 | 73 | All SZGR 2.0 genes in this pathway |