Gene Page: KDM5B
Summary ?
| GeneID | 10765 |
| Symbol | KDM5B |
| Synonyms | CT31|JARID1B|PLU-1|PLU1|PPP1R98|PUT1|RBBP2H1A|RBP2-H1 |
| Description | lysine demethylase 5B |
| Reference | MIM:605393|HGNC:HGNC:18039|Ensembl:ENSG00000117139|HPRD:09251|Vega:OTTHUMG00000041401 |
| Gene type | protein-coding |
| Map location | 1q32.1 |
| Sherlock p-value | 0.008 |
| Fetal beta | 2.034 |
| eGene | Cerebellum |
| Support | Ascano FMRP targets Chromatin Remodeling Genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
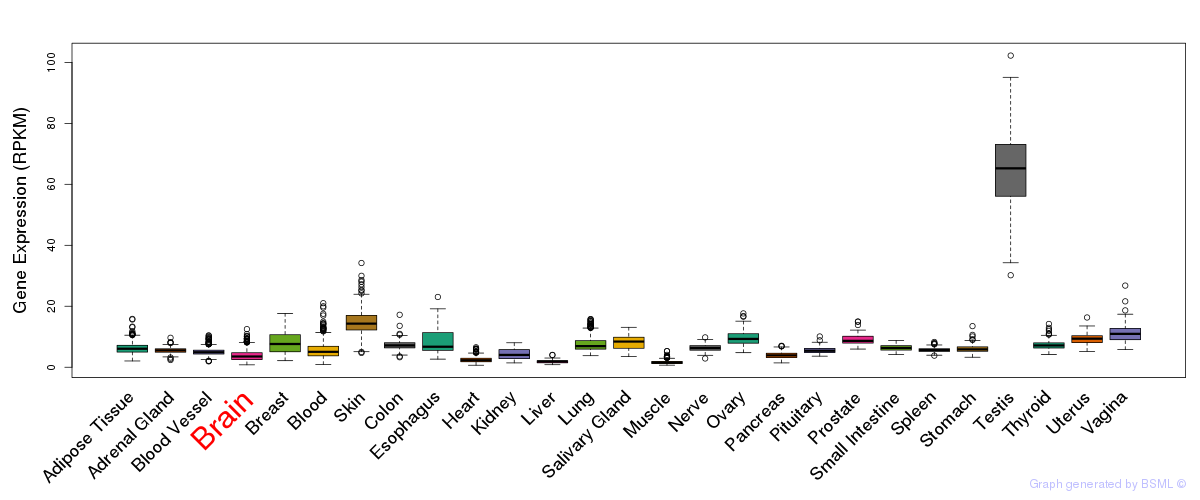
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PSMC6 | 0.90 | 0.92 |
| TXNDC9 | 0.88 | 0.89 |
| ACTR6 | 0.88 | 0.86 |
| TXNL1 | 0.87 | 0.84 |
| RSL24D1 | 0.87 | 0.85 |
| VBP1 | 0.86 | 0.83 |
| MRPL1 | 0.86 | 0.89 |
| COPS2 | 0.86 | 0.82 |
| YEATS4 | 0.86 | 0.83 |
| OLA1 | 0.85 | 0.82 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FAM38A | -0.58 | -0.61 |
| TENC1 | -0.58 | -0.59 |
| AF347015.26 | -0.56 | -0.56 |
| AF347015.2 | -0.56 | -0.55 |
| MICALL2 | -0.55 | -0.57 |
| AF347015.15 | -0.55 | -0.55 |
| GPT | -0.54 | -0.56 |
| AF347015.33 | -0.54 | -0.54 |
| AF347015.8 | -0.53 | -0.52 |
| MT-CYB | -0.52 | -0.53 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA UP | 305 | 185 | All SZGR 2.0 genes in this pathway |
| RODRIGUES DCC TARGETS DN | 121 | 84 | All SZGR 2.0 genes in this pathway |
| SMIRNOV CIRCULATING ENDOTHELIOCYTES IN CANCER UP | 158 | 103 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL DN | 275 | 168 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 DN | 281 | 186 | All SZGR 2.0 genes in this pathway |
| DITTMER PTHLH TARGETS UP | 112 | 68 | All SZGR 2.0 genes in this pathway |
| SCIBETTA KDM5B TARGETS DN | 81 | 55 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL UP | 316 | 190 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 CHRONIC LOF DN | 118 | 78 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| STREICHER LSM1 TARGETS UP | 44 | 34 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| GARCIA TARGETS OF FLI1 AND DAX1 UP | 57 | 34 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| PETRETTO BLOOD PRESSURE DN | 7 | 6 | All SZGR 2.0 genes in this pathway |
| XU GH1 AUTOCRINE TARGETS DN | 142 | 94 | All SZGR 2.0 genes in this pathway |
| DURCHDEWALD SKIN CARCINOGENESIS DN | 264 | 168 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY OVERCONNECTED IN BREAST CANCER | 22 | 19 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS UP | 317 | 208 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K9ME3 DN | 120 | 71 | All SZGR 2.0 genes in this pathway |
| IZADPANAH STEM CELL ADIPOSE VS BONE DN | 108 | 68 | All SZGR 2.0 genes in this pathway |
| RUIZ TNC TARGETS UP | 153 | 107 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE DN | 209 | 137 | All SZGR 2.0 genes in this pathway |
| CHANG CYCLING GENES | 148 | 83 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G1 UP | 113 | 70 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA UP | 207 | 143 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE M G1 | 148 | 95 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR DN | 505 | 328 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 4 UP | 112 | 64 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 7 UP | 118 | 68 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION UP | 271 | 165 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
| ZWANG EGF INTERVAL DN | 214 | 124 | All SZGR 2.0 genes in this pathway |