Gene Page: FOS
Summary ?
| GeneID | 2353 |
| Symbol | FOS |
| Synonyms | AP-1|C-FOS|p55 |
| Description | FBJ murine osteosarcoma viral oncogene homolog |
| Reference | MIM:164810|HGNC:HGNC:3796|Ensembl:ENSG00000170345|HPRD:01275|Vega:OTTHUMG00000171774 |
| Gene type | protein-coding |
| Map location | 14q24.3 |
| Pascal p-value | 0.327 |
| Sherlock p-value | 0.666 |
| Fetal beta | -1.196 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizotypy,schizophrenias,schizotypal | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg12110634 | 14 | 75725749 | FOS | 3.96E-9 | -0.01 | 2.45E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2591089 | chr14 | 76587799 | FOS | 2353 | 0.12 | cis | ||
| rs2543359 | chr14 | 76590472 | FOS | 2353 | 0.01 | cis | ||
| rs935340 | chr14 | 76591205 | FOS | 2353 | 0.02 | cis | ||
| rs12059674 | chr1 | 57630277 | FOS | 2353 | 5.909E-4 | trans | ||
| rs17126801 | chr1 | 65098162 | FOS | 2353 | 0.1 | trans | ||
| rs7511937 | 0 | FOS | 2353 | 0.04 | trans | |||
| rs17512695 | chr1 | 73988565 | FOS | 2353 | 0.16 | trans | ||
| rs17100274 | chr1 | 77774412 | FOS | 2353 | 0.03 | trans | ||
| rs6659821 | chr1 | 110339827 | FOS | 2353 | 0.04 | trans | ||
| rs4457633 | chr1 | 204283819 | FOS | 2353 | 0.19 | trans | ||
| rs1796829 | chr1 | 239661818 | FOS | 2353 | 0.04 | trans | ||
| rs12034521 | chr1 | 239717679 | FOS | 2353 | 0.01 | trans | ||
| snp_a-1990354 | 0 | FOS | 2353 | 5.02E-4 | trans | |||
| rs17038084 | chr2 | 85192280 | FOS | 2353 | 0 | trans | ||
| rs7574684 | chr2 | 100992911 | FOS | 2353 | 0.18 | trans | ||
| rs17024129 | chr2 | 100995144 | FOS | 2353 | 0.18 | trans | ||
| rs7585898 | chr2 | 100998292 | FOS | 2353 | 0.18 | trans | ||
| rs1370627 | chr2 | 101009384 | FOS | 2353 | 0.18 | trans | ||
| rs6707117 | chr2 | 136855504 | FOS | 2353 | 8.315E-7 | trans | ||
| rs10189091 | chr2 | 143471999 | FOS | 2353 | 0.03 | trans | ||
| rs16838911 | chr2 | 156087569 | FOS | 2353 | 0.04 | trans | ||
| rs16839796 | chr4 | 7188348 | FOS | 2353 | 0.03 | trans | ||
| rs4631023 | chr4 | 8101794 | FOS | 2353 | 0.13 | trans | ||
| rs6827713 | chr4 | 79936877 | FOS | 2353 | 0.11 | trans | ||
| rs10079220 | chr5 | 57453717 | FOS | 2353 | 0.1 | trans | ||
| rs10040715 | chr5 | 57594207 | FOS | 2353 | 6.951E-5 | trans | ||
| rs16894149 | chr5 | 61013724 | FOS | 2353 | 0.12 | trans | ||
| snp_a-4217300 | 0 | FOS | 2353 | 4.727E-4 | trans | |||
| rs163953 | chr5 | 92289536 | FOS | 2353 | 0.09 | trans | ||
| rs17134275 | chr5 | 100634998 | FOS | 2353 | 0.08 | trans | ||
| rs2141482 | chr5 | 136760477 | FOS | 2353 | 0.11 | trans | ||
| rs2189803 | chr5 | 137684661 | FOS | 2353 | 1.233E-5 | trans | ||
| rs12659857 | chr5 | 168306133 | FOS | 2353 | 0.2 | trans | ||
| snp_a-2055204 | 0 | FOS | 2353 | 0.03 | trans | |||
| rs259913 | chr5 | 169370782 | FOS | 2353 | 0.1 | trans | ||
| rs11965040 | chr6 | 18644344 | FOS | 2353 | 0.06 | trans | ||
| rs10945911 | chr6 | 164047509 | FOS | 2353 | 4.675E-4 | trans | ||
| rs12540348 | chr7 | 25802146 | FOS | 2353 | 0.16 | trans | ||
| rs4719825 | chr7 | 25813917 | FOS | 2353 | 0.01 | trans | ||
| rs12701006 | chr7 | 30147298 | FOS | 2353 | 0.13 | trans | ||
| rs1419793 | chr7 | 34697630 | FOS | 2353 | 0.14 | trans | ||
| rs10245449 | chr7 | 80935100 | FOS | 2353 | 0.08 | trans | ||
| rs10487383 | chr7 | 117671737 | FOS | 2353 | 0.02 | trans | ||
| rs1457044 | chr8 | 59257567 | FOS | 2353 | 0.14 | trans | ||
| rs1733950 | chr8 | 90062516 | FOS | 2353 | 5.245E-4 | trans | ||
| rs1355060 | chr8 | 118717710 | FOS | 2353 | 0.18 | trans | ||
| rs2280828 | chr8 | 118762435 | FOS | 2353 | 0.1 | trans | ||
| rs9298651 | chr9 | 10076275 | FOS | 2353 | 0 | trans | ||
| rs16931018 | chr9 | 10077048 | FOS | 2353 | 0 | trans | ||
| rs11791891 | chr9 | 87512411 | FOS | 2353 | 0.09 | trans | ||
| rs2165893 | chr9 | 87531423 | FOS | 2353 | 0.03 | trans | ||
| rs3780642 | chr9 | 87553144 | FOS | 2353 | 0.03 | trans | ||
| rs6537944 | chr9 | 137312827 | FOS | 2353 | 0.1 | trans | ||
| rs1908351 | chr10 | 57877117 | FOS | 2353 | 0.04 | trans | ||
| rs3104871 | chr10 | 57878502 | FOS | 2353 | 0 | trans | ||
| rs2856339 | chr12 | 11906818 | FOS | 2353 | 0.18 | trans | ||
| rs8490 | chr12 | 51453904 | FOS | 2353 | 2.568E-4 | trans | ||
| rs1344796 | chr12 | 76397104 | FOS | 2353 | 0.09 | trans | ||
| rs7133714 | chr12 | 120141546 | FOS | 2353 | 0.05 | trans | ||
| rs11833075 | chr12 | 122114592 | FOS | 2353 | 0.18 | trans | ||
| rs3803214 | chr13 | 25740255 | FOS | 2353 | 0.07 | trans | ||
| rs7982110 | chr13 | 59723428 | FOS | 2353 | 0.02 | trans | ||
| rs149642 | chr14 | 72653498 | FOS | 2353 | 0.14 | trans | ||
| rs4905035 | chr14 | 93489262 | FOS | 2353 | 0.03 | trans | ||
| rs1885073 | chr14 | 104357181 | FOS | 2353 | 0.01 | trans | ||
| rs8047151 | chr16 | 17209696 | FOS | 2353 | 0.02 | trans | ||
| rs4271578 | chr16 | 17213886 | FOS | 2353 | 0.09 | trans | ||
| rs7219986 | chr17 | 53263206 | FOS | 2353 | 0.2 | trans | ||
| rs9646596 | chr18 | 53049211 | FOS | 2353 | 0.13 | trans | ||
| rs11152369 | chr18 | 53066327 | FOS | 2353 | 0.18 | trans | ||
| rs1942464 | chr18 | 70509877 | FOS | 2353 | 0.04 | trans | ||
| rs1797040 | chr20 | 23907743 | FOS | 2353 | 0.08 | trans | ||
| rs2424603 | 0 | FOS | 2353 | 0.08 | trans | |||
| rs6102879 | chr20 | 41138766 | FOS | 2353 | 0.04 | trans | ||
| rs6026122 | chr20 | 56769083 | FOS | 2353 | 0.04 | trans | ||
| rs6015452 | chr20 | 57782312 | FOS | 2353 | 0.19 | trans | ||
| rs2834872 | chr21 | 36634845 | FOS | 2353 | 0.04 | trans | ||
| rs2834875 | chr21 | 36656119 | FOS | 2353 | 0.02 | trans | ||
| rs877910 | chr21 | 36658376 | FOS | 2353 | 0.12 | trans | ||
| rs134028 | chr22 | 28048448 | FOS | 2353 | 0.1 | trans | ||
| rs5768165 | chr22 | 48318197 | FOS | 2353 | 0.03 | trans | ||
| rs5768168 | chr22 | 48319334 | FOS | 2353 | 0.03 | trans | ||
| rs5768173 | chr22 | 48321475 | FOS | 2353 | 0.03 | trans | ||
| rs16978981 | chrX | 13538144 | FOS | 2353 | 0.03 | trans | ||
| rs7879159 | chrX | 19481591 | FOS | 2353 | 0.08 | trans | ||
| rs12006821 | chrX | 50424687 | FOS | 2353 | 0.03 | trans |
Section II. Transcriptome annotation
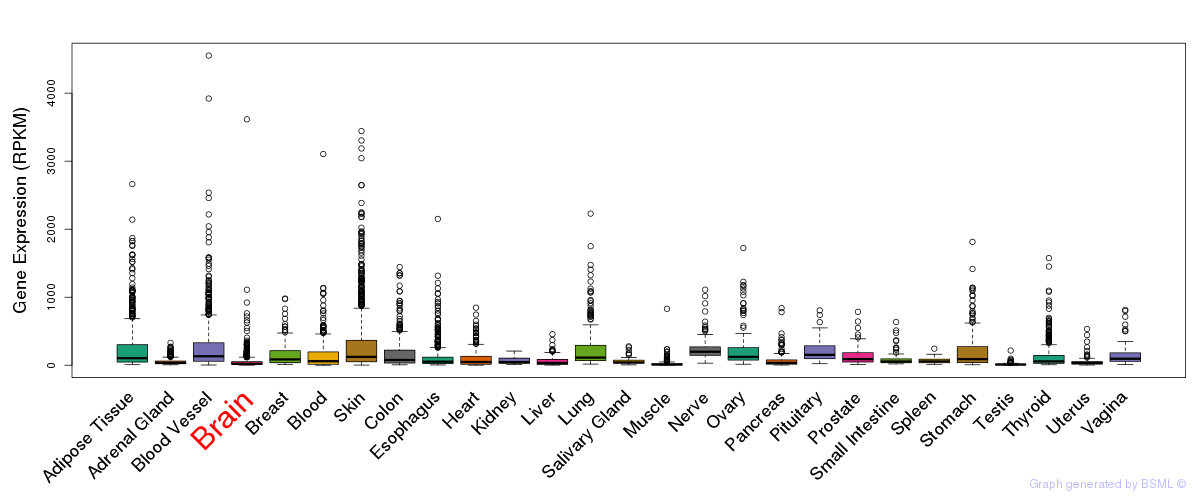
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | IEA | - | |
| GO:0003704 | specific RNA polymerase II transcription factor activity | TAS | 10918580 | |
| GO:0043565 | sequence-specific DNA binding | IEA | - | |
| GO:0046983 | protein dimerization activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007399 | nervous system development | IEA | neurite (GO term level: 5) | - |
| GO:0006306 | DNA methylation | TAS | 9888853 | |
| GO:0006357 | regulation of transcription from RNA polymerase II promoter | TAS | 10918580 | |
| GO:0006954 | inflammatory response | TAS | 9443941 | |
| GO:0034614 | cellular response to reactive oxygen species | IDA | 17217916 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | TAS | 9443941 | |
| GO:0005667 | transcription factor complex | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ATF2 | CRE-BP1 | CREB2 | HB16 | MGC111558 | TREB7 | activating transcription factor 2 | - | HPRD,BioGRID | 1828107 |
| BATF | B-ATF | BATF1 | SFA-2 | basic leucine zipper transcription factor, ATF-like | - | HPRD,BioGRID | 8570175 |
| BCL3 | BCL4 | D19S37 | B-cell CLL/lymphoma 3 | - | HPRD,BioGRID | 10497212 |
| CEBPG | GPE1BP | IG/EBP-1 | CCAAT/enhancer binding protein (C/EBP), gamma | - | HPRD | 7665092 |
| COBRA1 | DKFZp586B0519 | KIAA1182 | NELF-B | NELFB | cofactor of BRCA1 | - | HPRD,BioGRID | 15530430 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | Reconstituted Complex | BioGRID | 11782371 |
| CSNK2A1 | CK2A1 | CKII | casein kinase 2, alpha 1 polypeptide | - | HPRD,BioGRID | 9685505 |
| CSNK2A2 | CK2A2 | CSNK2A1 | FLJ43934 | casein kinase 2, alpha prime polypeptide | - | HPRD,BioGRID | 9685505 |
| DDIT3 | CEBPZ | CHOP | CHOP10 | GADD153 | MGC4154 | DNA-damage-inducible transcript 3 | - | HPRD,BioGRID | 10523647 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | An unspecified isoform of EGFR binds to c-fos promoter. | BIND | 15950906 |
| ELK1 | - | ELK1, member of ETS oncogene family | - | HPRD | 7540136 |
| ELK4 | SAP1 | ELK4, ETS-domain protein (SRF accessory protein 1) | - | HPRD | 7540136 |
| ELK4 | SAP1 | ELK4, ETS-domain protein (SRF accessory protein 1) | Sap-1a is interacts with the serum response element (SRE) of c-fos. This interaction was modeled on a demonstrated interaction between human Sap-1a and c-fos from an unspecified species. | BIND | 9130707 |
| ERG | erg-3 | p55 | v-ets erythroblastosis virus E26 oncogene homolog (avian) | - | HPRD | 9681824 |
| ETS1 | ETS-1 | EWSR2 | FLJ10768 | v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) | - | HPRD | 8557686 |
| ETS2 | ETS2IT1 | v-ets erythroblastosis virus E26 oncogene homolog 2 (avian) | Reconstituted Complex | BioGRID | 9334186 |
| GATA4 | MGC126629 | GATA binding protein 4 | - | HPRD | 9207128 |
| GTF2E2 | FE | TF2E2 | TFIIE-B | general transcription factor IIE, polypeptide 2, beta 34kDa | Reconstituted Complex | BioGRID | 8628277 |
| GTF2F1 | BTF4 | RAP74 | TF2F1 | TFIIF | general transcription factor IIF, polypeptide 1, 74kDa | Reconstituted Complex | BioGRID | 8628277 |
| GTF2F2 | BTF4 | RAP30 | TF2F2 | TFIIF | general transcription factor IIF, polypeptide 2, 30kDa | - | HPRD,BioGRID | 8628277 |
| HNF1A | HNF1 | LFB1 | MODY3 | TCF1 | HNF1 homeobox A | - | HPRD,BioGRID | 11134330 |
| JAG1 | AGS | AHD | AWS | CD339 | HJ1 | JAGL1 | MGC104644 | jagged 1 (Alagille syndrome) | c-Fos interacts with Jagged1 promoter. | BIND | 16023595 |
| JUN | AP-1 | AP1 | c-Jun | jun oncogene | c-Jun interacts with c-Fos. This interaction was modelled on a demonstrated interaction between c-Jun from an unspecified species and c-Fos from an unspecified species. | BIND | 10488148 |
| JUN | AP-1 | AP1 | c-Jun | jun oncogene | Co-crystal Structure Reconstituted Complex Two-hybrid | BioGRID | 7816143 |8380166 |8440710 |9160889 |9872330 |10488148 |11053448 |
| JUN | AP-1 | AP1 | c-Jun | jun oncogene | cFos interacts with cJun. | BIND | 8816797 |8816798 |
| JUN | AP-1 | AP1 | c-Jun | jun oncogene | - | HPRD | 1631061 |7816143 |9346889 |
| JUNB | AP-1 | jun B proto-oncogene | - | HPRD | 1406655|10764760 |
| JUND | AP-1 | jun D proto-oncogene | - | HPRD | 12193410 |
| KHSRP | FBP2 | FUBP2 | KSRP | MGC99676 | KH-type splicing regulatory protein | KSRP interacts with AREfos. This interaction was modeled on a demonstrated interaction between KSRP from human and AREfos from an unspecified species. | BIND | 15175153 |
| MAF | MGC71685 | c-MAF | v-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian) | - | HPRD | 8108109 |
| MAP3K7 | TAK1 | TGF1a | mitogen-activated protein kinase kinase kinase 7 | - | HPRD | 11397816 |
| MITF | MI | WS2A | bHLHe32 | microphthalmia-associated transcription factor | - | HPRD,BioGRID | 9918847 |
| NACA | HSD48 | MGC117224 | NACA1 | nascent polypeptide-associated complex alpha subunit | Reconstituted Complex | BioGRID | 9488446 |
| NCOA1 | F-SRC-1 | KAT13A | MGC129719 | MGC129720 | NCoA-1 | RIP160 | SRC-1 | SRC1 | bHLHe42 | nuclear receptor coactivator 1 | - | HPRD,BioGRID | 9642216 |
| NCOA6 | AIB3 | ANTP | ASC2 | HOX1.1 | HOXA7 | KIAA0181 | NRC | PRIP | RAP250 | TRBP | nuclear receptor coactivator 6 | Reconstituted Complex | BioGRID | 10847592 |
| NCOR2 | CTG26 | SMRT | SMRTE | SMRTE-tau | TNRC14 | TRAC-1 | TRAC1 | nuclear receptor co-repressor 2 | - | HPRD,BioGRID | 10777532 |
| NFATC2 | KIAA0611 | NFAT1 | NFATP | nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 | - | HPRD | 8397339 |12949493 |
| NFKB1 | DKFZp686C01211 | EBP-1 | KBF1 | MGC54151 | NF-kappa-B | NFKB-p105 | NFKB-p50 | p105 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 | - | HPRD | 9468519 |
| PML | MYL | PP8675 | RNF71 | TRIM19 | promyelocytic leukemia | - | HPRD,BioGRID | 9671405 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | - | HPRD | 8475068 |
| RELA | MGC131774 | NFKB3 | p65 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | NF-kappa-B interacts with c-Fos. | BIND | 10488148 |
| RELA | MGC131774 | NFKB3 | p65 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | Phenotypic Enhancement Two-hybrid | BioGRID | 10488148 |
| RPS6KA4 | MSK2 | RSK-B | ribosomal protein S6 kinase, 90kDa, polypeptide 4 | - | HPRD,BioGRID | 9792677 |
| RUNX1 | AML1 | AML1-EVI-1 | AMLCR1 | CBFA2 | EVI-1 | PEBP2aB | runt-related transcription factor 1 | - | HPRD,BioGRID | 11641401 |
| RUNX2 | AML3 | CBFA1 | CCD | CCD1 | MGC120022 | MGC120023 | OSF2 | PEA2aA | PEBP2A1 | PEBP2A2 | PEBP2aA | PEBP2aA1 | runt-related transcription factor 2 | - | HPRD,BioGRID | 11641401 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | Reconstituted Complex Two-hybrid | BioGRID | 9732876 |
| SMARCD1 | BAF60A | CRACD1 | Rsc6p | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 | - | HPRD | 11053448 |
| SPI1 | OF | PU.1 | SFPI1 | SPI-1 | SPI-A | spleen focus forming virus (SFFV) proviral integration oncogene spi1 | Reconstituted Complex | BioGRID | 9681824 |
| SRF | MCM1 | serum response factor (c-fos serum response element-binding transcription factor) | SRF interacts with the c-fos promoter CArG box. This interaction was modeled on a demonstrated interaction between human SRF and c-fos promoter CArG box from an unspecified species. | BIND | 15610731 |
| STAT3 | APRF | FLJ20882 | HIES | MGC16063 | signal transducer and activator of transcription 3 (acute-phase response factor) | An unspecified isoform of STAT3 binds to c-fos promoter. | BIND | 15950906 |
| TAF1 | BA2R | CCG1 | CCGS | DYT3 | KAT4 | N-TAF1 | NSCL2 | OF | P250 | TAF2A | TAFII250 | TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa | - | HPRD,BioGRID | 8065335 |
| TBP | GTF2D | GTF2D1 | MGC117320 | MGC126054 | MGC126055 | SCA17 | TFIID | TATA box binding protein | Affinity Capture-Western Reconstituted Complex | BioGRID | 8065335 |
| TBP | GTF2D | GTF2D1 | MGC117320 | MGC126054 | MGC126055 | SCA17 | TFIID | TATA box binding protein | - | HPRD | 7685215 |
| TSC22D3 | DIP | DKFZp313A1123 | DSIPI | GILZ | TSC-22R | hDIP | TSC22 domain family, member 3 | - | HPRD,BioGRID | 11397794 |
| USF2 | FIP | bHLHb12 | upstream transcription factor 2, c-fos interacting | - | HPRD | 1589769 |
| VDR | NR1I1 | vitamin D (1,25- dihydroxyvitamin D3) receptor | Reconstituted Complex | BioGRID | 10330159 |
| XBP1 | TREB5 | XBP2 | X-box binding protein 1 | - | HPRD,BioGRID | 1903538 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG TOLL LIKE RECEPTOR SIGNALING PATHWAY | 102 | 88 | All SZGR 2.0 genes in this pathway |
| KEGG T CELL RECEPTOR SIGNALING PATHWAY | 108 | 89 | All SZGR 2.0 genes in this pathway |
| KEGG B CELL RECEPTOR SIGNALING PATHWAY | 75 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG LEISHMANIA INFECTION | 72 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG COLORECTAL CANCER | 62 | 47 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BCELLSURVIVAL PATHWAY | 16 | 10 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BCR PATHWAY | 37 | 28 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RANKL PATHWAY | 14 | 13 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CDMAC PATHWAY | 16 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EGF PATHWAY | 31 | 25 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EPO PATHWAY | 19 | 13 | All SZGR 2.0 genes in this pathway |
| BIOCARTA FCER1 PATHWAY | 41 | 30 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IGF1 PATHWAY | 21 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL2 PATHWAY | 22 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL3 PATHWAY | 15 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL6 PATHWAY | 22 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL2RB PATHWAY | 38 | 26 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GLEEVEC PATHWAY | 23 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA INSULIN PATHWAY | 22 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA KERATINOCYTE PATHWAY | 46 | 38 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MAPK PATHWAY | 87 | 68 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ETS PATHWAY | 18 | 12 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NGF PATHWAY | 18 | 13 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ARENRF2 PATHWAY | 13 | 9 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PDGF PATHWAY | 32 | 25 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CCR5 PATHWAY | 20 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA DREAM PATHWAY | 14 | 13 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CARDIACEGF PATHWAY | 18 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MET PATHWAY | 37 | 30 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GPCR PATHWAY | 37 | 31 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TCR PATHWAY | 49 | 37 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TOLL PATHWAY | 37 | 31 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TPO PATHWAY | 24 | 17 | All SZGR 2.0 genes in this pathway |
| SA B CELL RECEPTOR COMPLEXES | 24 | 20 | All SZGR 2.0 genes in this pathway |
| ST GRANULE CELL SURVIVAL PATHWAY | 27 | 23 | All SZGR 2.0 genes in this pathway |
| PID SMAD2 3NUCLEAR PATHWAY | 82 | 63 | All SZGR 2.0 genes in this pathway |
| PID FCER1 PATHWAY | 62 | 43 | All SZGR 2.0 genes in this pathway |
| PID ENDOTHELIN PATHWAY | 63 | 52 | All SZGR 2.0 genes in this pathway |
| PID BCR 5PATHWAY | 65 | 50 | All SZGR 2.0 genes in this pathway |
| PID RHOA PATHWAY | 45 | 33 | All SZGR 2.0 genes in this pathway |
| PID LYSOPHOSPHOLIPID PATHWAY | 66 | 53 | All SZGR 2.0 genes in this pathway |
| PID GMCSF PATHWAY | 37 | 31 | All SZGR 2.0 genes in this pathway |
| PID IL12 2PATHWAY | 63 | 54 | All SZGR 2.0 genes in this pathway |
| PID NFAT TFPATHWAY | 47 | 39 | All SZGR 2.0 genes in this pathway |
| PID AVB3 OPN PATHWAY | 31 | 29 | All SZGR 2.0 genes in this pathway |
| PID PS1 PATHWAY | 46 | 39 | All SZGR 2.0 genes in this pathway |
| PID TELOMERASE PATHWAY | 68 | 48 | All SZGR 2.0 genes in this pathway |
| PID REG GR PATHWAY | 82 | 60 | All SZGR 2.0 genes in this pathway |
| PID IL2 1PATHWAY | 55 | 43 | All SZGR 2.0 genes in this pathway |
| PID TCR RAS PATHWAY | 14 | 14 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 DOWNSTREAM PATHWAY | 105 | 78 | All SZGR 2.0 genes in this pathway |
| PID ATF2 PATHWAY | 59 | 43 | All SZGR 2.0 genes in this pathway |
| PID AP1 PATHWAY | 70 | 60 | All SZGR 2.0 genes in this pathway |
| PID ERBB2 ERBB3 PATHWAY | 44 | 35 | All SZGR 2.0 genes in this pathway |
| PID FOXM1 PATHWAY | 40 | 30 | All SZGR 2.0 genes in this pathway |
| PID IL6 7 PATHWAY | 47 | 40 | All SZGR 2.0 genes in this pathway |
| PID PDGFRB PATHWAY | 129 | 103 | All SZGR 2.0 genes in this pathway |
| PID PDGFRA PATHWAY | 22 | 18 | All SZGR 2.0 genes in this pathway |
| PID AR NONGENOMIC PATHWAY | 31 | 27 | All SZGR 2.0 genes in this pathway |
| PID TCR CALCIUM PATHWAY | 29 | 23 | All SZGR 2.0 genes in this pathway |
| PID HIF1 TFPATHWAY | 66 | 52 | All SZGR 2.0 genes in this pathway |
| PID S1P S1P2 PATHWAY | 24 | 19 | All SZGR 2.0 genes in this pathway |
| PID MAPK TRK PATHWAY | 34 | 31 | All SZGR 2.0 genes in this pathway |
| PID CD8 TCR DOWNSTREAM PATHWAY | 65 | 56 | All SZGR 2.0 genes in this pathway |
| PID FGF PATHWAY | 55 | 37 | All SZGR 2.0 genes in this pathway |
| PID HNF3A PATHWAY | 44 | 29 | All SZGR 2.0 genes in this pathway |
| PID IL12 STAT4 PATHWAY | 33 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME TRIF MEDIATED TLR3 SIGNALING | 74 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME MAP KINASE ACTIVATION IN TLR CASCADE | 50 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | 10 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME MAPK TARGETS NUCLEAR EVENTS MEDIATED BY MAP KINASES | 30 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAF6 MEDIATED INDUCTION OF NFKB AND MAP KINASES UPON TLR7 8 OR 9 ACTIVATION | 77 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME NFKB AND MAP KINASES ACTIVATION MEDIATED BY TLR4 SIGNALING REPERTOIRE | 72 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | 83 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATED TLR4 SIGNALLING | 93 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME TOLL RECEPTOR CASCADES | 118 | 84 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| SOTIRIOU BREAST CANCER GRADE 1 VS 3 DN | 52 | 34 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST DUCTAL CARCINOMA VS DUCTAL NORMAL DN | 198 | 110 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST DUCTAL CARCINOMA VS LOBULAR NORMAL DN | 69 | 43 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS TOP50 DN | 45 | 26 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE DN | 384 | 220 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA UP | 177 | 110 | All SZGR 2.0 genes in this pathway |
| CORRE MULTIPLE MYELOMA DN | 62 | 41 | All SZGR 2.0 genes in this pathway |
| CASORELLI APL SECONDARY VS DE NOVO UP | 39 | 25 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS DN | 384 | 230 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL UP | 233 | 161 | All SZGR 2.0 genes in this pathway |
| BILBAN B CLL LPL DN | 42 | 25 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY DN | 362 | 238 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS DN | 260 | 143 | All SZGR 2.0 genes in this pathway |
| LEE NEURAL CREST STEM CELL DN | 118 | 79 | All SZGR 2.0 genes in this pathway |
| NAGASHIMA NRG1 SIGNALING UP | 176 | 123 | All SZGR 2.0 genes in this pathway |
| NAGASHIMA EGF SIGNALING UP | 58 | 40 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA UP | 171 | 112 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA BY DMOG UP | 130 | 85 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HIF1A TARGETS DN | 91 | 58 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HIF1A AND HIF2A TARGETS DN | 104 | 72 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS UP | 149 | 84 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER DN | 232 | 154 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION KRAS CDC25 DN | 51 | 35 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION KRAS DN | 142 | 95 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN UP | 239 | 157 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS E UP | 97 | 60 | All SZGR 2.0 genes in this pathway |
| JOHANSSON GLIOMAGENESIS BY PDGFB UP | 58 | 44 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK UP | 271 | 175 | All SZGR 2.0 genes in this pathway |
| HU ANGIOGENESIS UP | 21 | 16 | All SZGR 2.0 genes in this pathway |
| CHASSOT SKIN WOUND | 10 | 9 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION 12HR | 43 | 35 | All SZGR 2.0 genes in this pathway |
| RODRIGUES NTN1 AND DCC TARGETS | 35 | 25 | All SZGR 2.0 genes in this pathway |
| TURJANSKI MAPK1 AND MAPK2 TARGETS | 12 | 10 | All SZGR 2.0 genes in this pathway |
| TURJANSKI MAPK7 TARGETS | 8 | 8 | All SZGR 2.0 genes in this pathway |
| TURJANSKI MAPK11 TARGETS | 5 | 5 | All SZGR 2.0 genes in this pathway |
| TURJANSKI MAPK14 TARGETS | 10 | 10 | All SZGR 2.0 genes in this pathway |
| HUMMERICH SKIN CANCER PROGRESSION UP | 88 | 58 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 1 | 45 | 27 | All SZGR 2.0 genes in this pathway |
| KOBAYASHI RESPONSE TO ROMIDEPSIN | 19 | 14 | All SZGR 2.0 genes in this pathway |
| DACOSTA ERCC3 ALLELE XPCS VS TTD UP | 28 | 19 | All SZGR 2.0 genes in this pathway |
| AMUNDSON GENOTOXIC SIGNATURE | 105 | 68 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS NEUROEPITHELIUM DN | 164 | 111 | All SZGR 2.0 genes in this pathway |
| BORLAK LIVER CANCER EGF UP | 57 | 41 | All SZGR 2.0 genes in this pathway |
| STANHILL HRAS TRANSFROMATION UP | 8 | 5 | All SZGR 2.0 genes in this pathway |
| KHETCHOUMIAN TRIM24 TARGETS UP | 47 | 38 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 DN | 156 | 106 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA HIF1A DN | 110 | 78 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 AND HIF1A UP | 142 | 104 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION DN | 206 | 136 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 20 HELA | 11 | 8 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 40 MCF10A | 19 | 11 | All SZGR 2.0 genes in this pathway |
| AMIT SERUM RESPONSE 40 MCF10A | 32 | 21 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| GOTTWEIN TARGETS OF KSHV MIR K12 11 | 63 | 45 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 DN | 464 | 276 | All SZGR 2.0 genes in this pathway |
| SHEPARD CRUSH AND BURN MUTANT UP | 197 | 110 | All SZGR 2.0 genes in this pathway |
| FRASOR RESPONSE TO ESTRADIOL UP | 37 | 28 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO UP | 205 | 126 | All SZGR 2.0 genes in this pathway |
| CHESLER BRAIN HIGHEST EXPRESSION | 40 | 29 | All SZGR 2.0 genes in this pathway |
| TENEDINI MEGAKARYOCYTE MARKERS | 66 | 48 | All SZGR 2.0 genes in this pathway |
| JECHLINGER EPITHELIAL TO MESENCHYMAL TRANSITION DN | 66 | 47 | All SZGR 2.0 genes in this pathway |
| IIZUKA LIVER CANCER PROGRESSION L1 G1 UP | 25 | 15 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| RADAEVA RESPONSE TO IFNA1 DN | 10 | 5 | All SZGR 2.0 genes in this pathway |
| NING CHRONIC OBSTRUCTIVE PULMONARY DISEASE UP | 157 | 105 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR DN | 25 | 15 | All SZGR 2.0 genes in this pathway |
| ZHANG TARGETS OF EWSR1 FLI1 FUSION | 88 | 68 | All SZGR 2.0 genes in this pathway |
| MA MYELOID DIFFERENTIATION DN | 44 | 30 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| ASTIER INTEGRIN SIGNALING | 59 | 44 | All SZGR 2.0 genes in this pathway |
| HESS TARGETS OF HOXA9 AND MEIS1 DN | 77 | 48 | All SZGR 2.0 genes in this pathway |
| NEMETH INFLAMMATORY RESPONSE LPS DN | 32 | 25 | All SZGR 2.0 genes in this pathway |
| GERY CEBP TARGETS | 126 | 90 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS UP | 214 | 133 | All SZGR 2.0 genes in this pathway |
| MUNSHI MULTIPLE MYELOMA UP | 81 | 52 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS RESPONSE | 35 | 28 | All SZGR 2.0 genes in this pathway |
| HARRIS HYPOXIA | 81 | 64 | All SZGR 2.0 genes in this pathway |
| MCDOWELL ACUTE LUNG INJURY UP | 45 | 29 | All SZGR 2.0 genes in this pathway |
| CHANG IMMORTALIZED BY HPV31 DN | 65 | 45 | All SZGR 2.0 genes in this pathway |
| CHIBA RESPONSE TO TSA UP | 52 | 33 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 8HR UP | 105 | 73 | All SZGR 2.0 genes in this pathway |
| MAHAJAN RESPONSE TO IL1A DN | 76 | 57 | All SZGR 2.0 genes in this pathway |
| INGA TP53 TARGETS | 17 | 13 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR UP | 156 | 101 | All SZGR 2.0 genes in this pathway |
| GAJATE RESPONSE TO TRABECTEDIN UP | 67 | 45 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK UP | 244 | 151 | All SZGR 2.0 genes in this pathway |
| LEE AGING NEOCORTEX UP | 89 | 59 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS PEAK AT 2HR | 51 | 36 | All SZGR 2.0 genes in this pathway |
| MCCLUNG CREB1 TARGETS UP | 100 | 72 | All SZGR 2.0 genes in this pathway |
| LEE AGING CEREBELLUM UP | 84 | 58 | All SZGR 2.0 genes in this pathway |
| MCCLUNG COCAIN REWARD 4WK | 75 | 47 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV SCC UP | 123 | 75 | All SZGR 2.0 genes in this pathway |
| LEE AGING MUSCLE UP | 45 | 33 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 9 | 92 | 59 | All SZGR 2.0 genes in this pathway |
| BILD HRAS ONCOGENIC SIGNATURE | 261 | 166 | All SZGR 2.0 genes in this pathway |
| DURCHDEWALD SKIN CARCINOGENESIS DN | 264 | 168 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE DN | 373 | 196 | All SZGR 2.0 genes in this pathway |
| HILLION HMGA1 TARGETS | 90 | 71 | All SZGR 2.0 genes in this pathway |
| HILLION HMGA1B TARGETS | 92 | 68 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P7 | 90 | 52 | All SZGR 2.0 genes in this pathway |
| WANG LSD1 TARGETS DN | 39 | 30 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN | 409 | 268 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| OUYANG PROSTATE CANCER PROGRESSION UP | 20 | 14 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| MISHRA CARCINOMA ASSOCIATED FIBROBLAST UP | 24 | 14 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE DN | 274 | 165 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| BHATI G2M ARREST BY 2METHOXYESTRADIOL UP | 125 | 68 | All SZGR 2.0 genes in this pathway |
| CHEN HOXA5 TARGETS 9HR UP | 223 | 132 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA METAGENE | 242 | 168 | All SZGR 2.0 genes in this pathway |
| KRISHNAN FURIN TARGETS UP | 12 | 6 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC | 420 | 269 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C D DN | 252 | 155 | All SZGR 2.0 genes in this pathway |
| SPIRA SMOKERS LUNG CANCER UP | 38 | 24 | All SZGR 2.0 genes in this pathway |
| ZAIDI OSTEOBLAST TRANSCRIPTION FACTORS | 14 | 12 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| SCHRAETS MLL TARGETS UP | 35 | 21 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR UP | 293 | 203 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| CROONQUIST STROMAL STIMULATION UP | 60 | 42 | All SZGR 2.0 genes in this pathway |
| CROONQUIST NRAS VS STROMAL STIMULATION DN | 99 | 65 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER HEPATOBLAST | 16 | 12 | All SZGR 2.0 genes in this pathway |
| ONO AML1 TARGETS UP | 24 | 14 | All SZGR 2.0 genes in this pathway |
| ONO FOXP3 TARGETS UP | 23 | 15 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS DN | 306 | 191 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 2 | 86 | 50 | All SZGR 2.0 genes in this pathway |
| MARIADASON RESPONSE TO BUTYRATE CURCUMIN SULINDAC TSA 1 | 10 | 7 | All SZGR 2.0 genes in this pathway |
| NIELSEN GIST | 98 | 66 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 30MIN UP | 56 | 38 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 2HR UP | 39 | 30 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 DN | 374 | 217 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS VIA AKT1 UP | 281 | 183 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| DALESSIO TSA RESPONSE | 29 | 16 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 5 | 11 | 8 | All SZGR 2.0 genes in this pathway |
| LIU IL13 PRIMING MODEL | 15 | 11 | All SZGR 2.0 genes in this pathway |
| PHONG TNF TARGETS UP | 63 | 43 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE VIA P38 COMPLETE | 227 | 151 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 3 TRANSIENTLY INDUCED BY EGF | 222 | 159 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-101 | 524 | 531 | 1A,m8 | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-139 | 526 | 532 | m8 | hsa-miR-139brain | UCUACAGUGCACGUGUCU |
| miR-142-5p | 483 | 489 | m8 | hsa-miR-142-5p | CAUAAAGUAGAAAGCACUAC |
| miR-144 | 525 | 531 | 1A | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-181 | 592 | 598 | m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-221/222 | 203 | 210 | 1A,m8 | hsa-miR-221brain | AGCUACAUUGUCUGCUGGGUUUC |
| hsa-miR-222brain | AGCUACAUCUGGCUACUGGGUCUC | ||||
| miR-29 | 365 | 371 | m8 | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
| miR-338 | 368 | 374 | 1A | hsa-miR-338brain | UCCAGCAUCAGUGAUUUUGUUGA |
| miR-383 | 567 | 573 | 1A | hsa-miR-383brain | AGAUCAGAAGGUGAUUGUGGCU |
| miR-543 | 593 | 599 | m8 | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.