Gene Page: GSK3B
Summary ?
| GeneID | 2932 |
| Symbol | GSK3B |
| Synonyms | - |
| Description | glycogen synthase kinase 3 beta |
| Reference | MIM:605004|HGNC:HGNC:4617|Ensembl:ENSG00000082701|HPRD:05418|Vega:OTTHUMG00000133765 |
| Gene type | protein-coding |
| Map location | 3q13.3 |
| Pascal p-value | 0.866 |
| Sherlock p-value | 0.682 |
| Fetal beta | 0.554 |
| eGene | Myers' cis & trans |
| Support | INTRACELLULAR SIGNAL TRANSDUCTION G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.humanNRC G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Darnell FMRP targets Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 2 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.4135 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7715946 | chr5 | 146925366 | GSK3B | 2932 | 0.14 | trans | ||
| rs16930362 | chr10 | 74890320 | GSK3B | 2932 | 0.09 | trans | ||
| rs16915029 | chr10 | 75001994 | GSK3B | 2932 | 0.14 | trans | ||
| rs16930466 | chr10 | 75007587 | GSK3B | 2932 | 0.16 | trans | ||
| rs16930573 | chr10 | 75167645 | GSK3B | 2932 | 0.16 | trans | ||
| rs12571454 | chr10 | 75208954 | GSK3B | 2932 | 0.05 | trans | ||
| rs1324669 | chr13 | 107872446 | GSK3B | 2932 | 0.06 | trans | ||
| rs16958358 | chr17 | 55502410 | GSK3B | 2932 | 0.09 | trans |
Section II. Transcriptome annotation
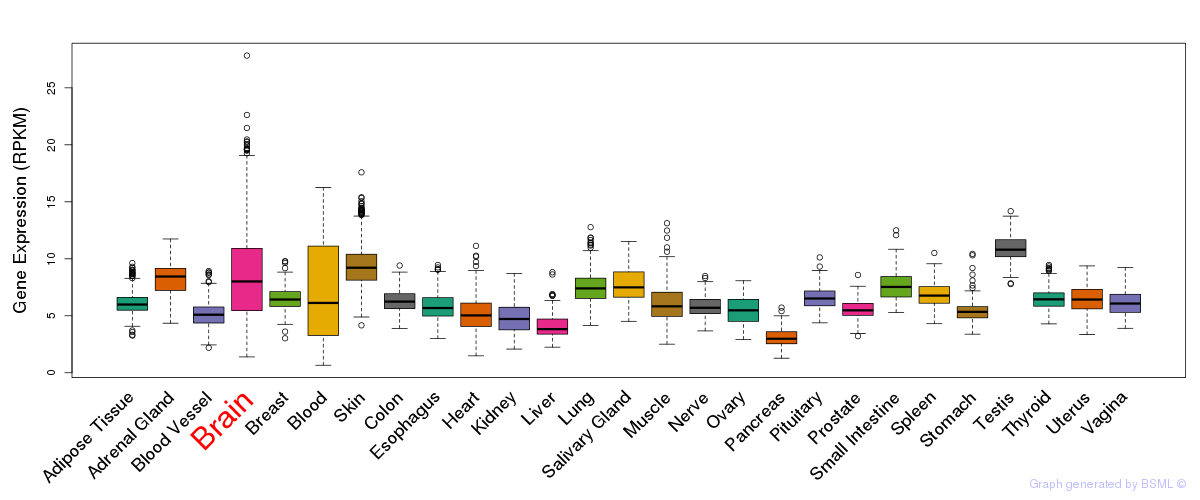
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SLC2A4 | 0.68 | 0.60 |
| SELENBP1 | 0.66 | 0.68 |
| FAM107A | 0.65 | 0.64 |
| ACADS | 0.65 | 0.68 |
| ALDH1L1 | 0.65 | 0.64 |
| CD40 | 0.65 | 0.58 |
| CRYL1 | 0.64 | 0.73 |
| ALDH2 | 0.63 | 0.51 |
| FAH | 0.62 | 0.67 |
| C10orf54 | 0.62 | 0.59 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| STRBP | -0.46 | -0.54 |
| ARIH1 | -0.46 | -0.56 |
| PURG | -0.45 | -0.51 |
| TMEM169 | -0.45 | -0.54 |
| GFPT1 | -0.45 | -0.53 |
| PAPD5 | -0.45 | -0.53 |
| LEO1 | -0.45 | -0.52 |
| RTF1 | -0.45 | -0.50 |
| FAM32A | -0.44 | -0.52 |
| NOVA2 | -0.44 | -0.52 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0050321 | tau-protein kinase activity | IEA | Brain (GO term level: 7) | - |
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0002039 | p53 binding | IDA | 14744935 | |
| GO:0004696 | glycogen synthase kinase 3 activity | IDA | 14744935 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0051059 | NF-kappaB binding | IPI | 15465828 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007242 | intracellular signaling cascade | IDA | 14749367 | |
| GO:0005977 | glycogen metabolic process | TAS | 7980435 | |
| GO:0006983 | ER overload response | IDA | 14744935 | |
| GO:0043066 | negative regulation of apoptosis | IDA | 14744935 | |
| GO:0018105 | peptidyl-serine phosphorylation | IDA | 11104755 |14744935 | |
| GO:0046827 | positive regulation of protein export from nucleus | IDA | 14744935 | |
| GO:0060070 | Wnt receptor signaling pathway through beta-catenin | IC | 9601641 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 8524413 |11955436 |12000790 |12820959 |15327769 |16753179 | |
| GO:0005634 | nucleus | IDA | 14744935 | |
| GO:0005737 | cytoplasm | IDA | 14744935 | |
| GO:0030877 | beta-catenin destruction complex | IDA | 9601641 |16188939 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACLY | ACL | ATPCL | CLATP | ATP citrate lyase | Affinity Capture-MS | BioGRID | 17353931 |
| AKAP11 | AKAP220 | DKFZp781I12161 | FLJ11304 | KIAA0629 | PRKA11 | A kinase (PRKA) anchor protein 11 | - | HPRD,BioGRID | 12147701 |
| AKT1 | AKT | MGC99656 | PKB | PKB-ALPHA | PRKBA | RAC | RAC-ALPHA | v-akt murine thymoma viral oncogene homolog 1 | AKT1 interacts with and phosphorylates GSK3B. This interaction was modelled on a demonstrated interaction between human AKT1 and GSK3B from an unspecified species. | BIND | 15688030 |
| AKT1 | AKT | MGC99656 | PKB | PKB-ALPHA | PRKBA | RAC | RAC-ALPHA | v-akt murine thymoma viral oncogene homolog 1 | Reconstituted Complex | BioGRID | 16009706 |
| AKT2 | PKBB | PKBBETA | PRKBB | RAC-BETA | v-akt murine thymoma viral oncogene homolog 2 | - | HPRD,BioGRID | 12434148 |
| APC | BTPS2 | DP2 | DP2.5 | DP3 | GS | adenomatous polyposis coli | Affinity Capture-Western | BioGRID | 11251183 |
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | Affinity Capture-Western Reconstituted Complex | BioGRID | 15178691 |
| AXIN1 | AXIN | MGC52315 | axin 1 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 9734785 |12511557 |
| AXIN2 | AXIL | DKFZp781B0869 | MGC10366 | MGC126582 | axin 2 | - | HPRD,BioGRID | 10966653 |
| BCL3 | BCL4 | D19S37 | B-cell CLL/lymphoma 3 | GSK3-beta interacts with and phosphorylates BCL-3. | BIND | 15469820 |
| BXDC1 | FLJ21087 | RPF2 | bA397G5.4 | brix domain containing 1 | Affinity Capture-MS | BioGRID | 17353931 |
| C14orf129 | GSKIP | HSPC210 | MGC4945 | chromosome 14 open reading frame 129 | Affinity Capture-MS | BioGRID | 17353931 |
| CCNE1 | CCNE | cyclin E1 | Biochemical Activity | BioGRID | 14536078 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | GSK3B (GSK3-Beta) interacts with and phosphorylates CTNNB1 (Beta-catenin). | BIND | 15829978 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | Affinity Capture-Western Biochemical Activity | BioGRID | 10330181 |11251183 |
| DDX20 | DKFZp434H052 | DP103 | GEMIN3 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 20 | Affinity Capture-MS | BioGRID | 17353931 |
| DHX36 | DDX36 | G4R1 | KIAA1488 | MLEL1 | RHAU | DEAH (Asp-Glu-Ala-His) box polypeptide 36 | Affinity Capture-MS | BioGRID | 17353931 |
| DNM1L | DLP1 | DRP1 | DVLP | DYMPLE | DYNIV-11 | FLJ41912 | HDYNIV | VPS1 | dynamin 1-like | - | HPRD,BioGRID | 9731200 |
| DVL1 | DVL | MGC54245 | dishevelled, dsh homolog 1 (Drosophila) | - | HPRD | 10428961 |
| EWSR1 | EWS | Ewing sarcoma breakpoint region 1 | Affinity Capture-MS | BioGRID | 17353931 |
| FRAT1 | FLJ97193 | frequently rearranged in advanced T-cell lymphomas | - | HPRD | 12095675 |
| FRAT2 | MGC10562 | frequently rearranged in advanced T-cell lymphomas 2 | - | HPRD | 11738041 |
| GEMIN4 | DKFZp434B131 | DKFZp434D174 | HC56 | HCAP1 | HHRF-1 | p97 | gem (nuclear organelle) associated protein 4 | Affinity Capture-MS | BioGRID | 17353931 |
| GSK3B | - | glycogen synthase kinase 3 beta | - | HPRD | 14529625 |
| IGF2BP1 | CRD-BP | CRDBP | IMP-1 | IMP1 | VICKZ1 | ZBP1 | insulin-like growth factor 2 mRNA binding protein 1 | Affinity Capture-MS | BioGRID | 17353931 |
| ILK | DKFZp686F1765 | P59 | integrin-linked kinase | - | HPRD | 9736715 |
| MAPT | DDPAC | FLJ31424 | FTDP-17 | MAPTL | MGC138549 | MSTD | MTBT1 | MTBT2 | PPND | TAU | microtubule-associated protein tau | GSK3beta phosphorylates Ftau.This interaction was modelled on a demonstrated interaction between rabbit GSK3beta and human Ftau. | BIND | 14636947 |
| MUC1 | CD227 | EMA | H23AG | MAM6 | PEM | PEMT | PUM | mucin 1, cell surface associated | - | HPRD,BioGRID | 9819408 |
| NCOA3 | ACTR | AIB-1 | AIB1 | CAGH16 | CTG26 | KAT13B | MGC141848 | RAC3 | SRC3 | TNRC14 | TNRC16 | TRAM-1 | pCIP | nuclear receptor coactivator 3 | SRC-3 interacts with and is phosphorylated by GSK3-beta. | BIND | 15383283 |
| NFKB1 | DKFZp686C01211 | EBP-1 | KBF1 | MGC54151 | NF-kappa-B | NFKB-p105 | NFKB-p50 | p105 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 | - | HPRD,BioGRID | 11425860 |
| NIN | KIAA1565 | ninein (GSK3B interacting protein) | - | HPRD,BioGRID | 11004522 |
| NOTCH1 | TAN1 | hN1 | Notch homolog 1, translocation-associated (Drosophila) | - | HPRD,BioGRID | 12123574 |
| NOTCH2 | AGS2 | hN2 | Notch homolog 2 (Drosophila) | - | HPRD,BioGRID | 12794074 |
| OGT | FLJ23071 | HRNT1 | MGC22921 | O-GLCNAC | O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase) | - | HPRD | 10753899 |
| PPP1CA | MGC15877 | MGC1674 | PP-1A | PPP1A | protein phosphatase 1, catalytic subunit, alpha isoform | Affinity Capture-Western | BioGRID | 12147701 |
| PPYR1 | MGC116897 | NPY4-R | NPY4R | PP1 | Y4 | pancreatic polypeptide receptor 1 | - | HPRD | 12147701 |
| PRKAR2A | MGC3606 | PKR2 | PRKAR2 | protein kinase, cAMP-dependent, regulatory, type II, alpha | Affinity Capture-Western | BioGRID | 12147701 |
| PRKCA | AAG6 | MGC129900 | MGC129901 | PKC-alpha | PKCA | PRKACA | protein kinase C, alpha | Biochemical Activity | BioGRID | 1324914 |
| PRKCB | MGC41878 | PKC-beta | PKCB | PRKCB1 | PRKCB2 | protein kinase C, beta | - | HPRD | 1324914 |
| PRKCD | MAY1 | MGC49908 | PKCD | nPKC-delta | protein kinase C, delta | PKC-delta interacts with and phosphorylates GSK3-beta. This interaction was modeled on a demonstrated interaction between human PKC-delta and rabbit GSK3-beta. | BIND | 15824731 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | - | HPRD,BioGRID | 11809746 |
| PTPN1 | PTP1B | protein tyrosine phosphatase, non-receptor type 1 | - | HPRD,BioGRID | 7514173 |
| SF3B1 | PRP10 | PRPF10 | SAP155 | SF3b155 | splicing factor 3b, subunit 1, 155kDa | Affinity Capture-MS | BioGRID | 17353931 |
| SGK3 | CISK | DKFZp781N0293 | SGK2 | SGKL | serum/glucocorticoid regulated kinase family, member 3 | - | HPRD,BioGRID | 12054501 |
| SMN2 | BCD541 | C-BCD541 | FLJ76644 | MGC20996 | MGC5208 | SMNC | survival of motor neuron 2, centromeric | Affinity Capture-MS | BioGRID | 17353931 |
| SMYD2 | HSKM-B | KMT3C | MGC119305 | ZMYND14 | SET and MYND domain containing 2 | Affinity Capture-MS | BioGRID | 17353931 |
| SNAI1 | SLUGH2 | SNA | SNAH | dJ710H13.1 | snail homolog 1 (Drosophila) | Snail interacts with GSK-3beta. | BIND | 15448698 |
| SNRNP70 | RNPU1Z | RPU1 | SNRP70 | U170K | U1AP | U1RNP | small nuclear ribonucleoprotein 70kDa (U1) | Affinity Capture-MS | BioGRID | 17353931 |
| STRAP | MAWD | PT-WD | UNRIP | serine/threonine kinase receptor associated protein | Affinity Capture-MS | BioGRID | 17353931 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | p53 interacts with GSK3-beta. | BIND | 12048243 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | - | HPRD,BioGRID | 12048243 |
| TPPP | TPPP/p25 | TPPP1 | p24 | p25 | p25alpha | tubulin polymerization promoting protein | - | HPRD | 11781156 |
| TSC1 | KIAA0243 | LAM | MGC86987 | TSC | tuberous sclerosis 1 | Affinity Capture-Western | BioGRID | 12511557 |
| TSC2 | FLJ43106 | LAM | TSC4 | tuberous sclerosis 2 | Affinity Capture-Western Biochemical Activity | BioGRID | 12511557 |16959574 |
| UPF3A | HUPF3A | RENT3A | UPF3 | UPF3 regulator of nonsense transcripts homolog A (yeast) | - | HPRD,BioGRID | 15231747 |
| XPO6 | EXP6 | FLJ22519 | KIAA0370 | RANBP20 | exportin 6 | Affinity Capture-MS | BioGRID | 17353931 |
Section V. Pathway annotation
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-199 | 61 | 67 | m8 | hsa-miR-199a | CCCAGUGUUCAGACUACCUGUUC |
| hsa-miR-199b | CCCAGUGUUUAGACUAUCUGUUC | ||||
| miR-26 | 41 | 47 | m8 | hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC |
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.