| KEGG SYSTEMIC LUPUS ERYTHEMATOSUS
| 140 | 100 | All SZGR 2.0 genes in this pathway |
| PID FANCONI PATHWAY
| 47 | 28 | All SZGR 2.0 genes in this pathway |
| PID ATM PATHWAY
| 34 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME MEIOSIS
| 116 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE
| 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I TRANSCRIPTION
| 89 | 64 | All SZGR 2.0 genes in this pathway |
| REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS
| 17 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTION
| 210 | 127 | All SZGR 2.0 genes in this pathway |
| REACTOME DOUBLE STRAND BREAK REPAIR
| 24 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION
| 122 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPAIR
| 112 | 59 | All SZGR 2.0 genes in this pathway |
| REACTOME CHROMOSOME MAINTENANCE
| 122 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE
| 64 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL I PROMOTER OPENING
| 62 | 49 | All SZGR 2.0 genes in this pathway |
| REACTOME MEIOTIC RECOMBINATION
| 86 | 62 | All SZGR 2.0 genes in this pathway |
| REACTOME MEIOTIC SYNAPSIS
| 73 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME AMYLOIDS
| 83 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME PACKAGING OF TELOMERE ENDS
| 48 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME TELOMERE MAINTENANCE
| 75 | 57 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP
| 783 | 507 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN
| 493 | 298 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP
| 579 | 346 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP
| 368 | 234 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS DN
| 310 | 188 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS UP
| 290 | 177 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS UP
| 372 | 227 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN
| 536 | 332 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML QUIESCENT VS NORMAL QUIESCENT UP
| 87 | 45 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML DIVIDING VS NORMAL QUIESCENT UP
| 181 | 101 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP
| 530 | 342 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP
| 443 | 294 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION KRAS UP
| 126 | 72 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION CDC25 UP
| 120 | 73 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF DN
| 228 | 137 | All SZGR 2.0 genes in this pathway |
| KONG E2F1 TARGETS
| 10 | 6 | All SZGR 2.0 genes in this pathway |
| KONG E2F3 TARGETS
| 97 | 58 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN
| 800 | 473 | All SZGR 2.0 genes in this pathway |
| EBAUER TARGETS OF PAX3 FOXO1 FUSION UP
| 207 | 128 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 4
| 61 | 34 | All SZGR 2.0 genes in this pathway |
| ROSTY CERVICAL CANCER PROLIFERATION CLUSTER
| 140 | 73 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER
| 747 | 453 | All SZGR 2.0 genes in this pathway |
| OXFORD RALA OR RALB TARGETS UP
| 48 | 23 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP
| 414 | 287 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP
| 479 | 299 | All SZGR 2.0 genes in this pathway |
| PUJANA BREAST CANCER LIT INT NETWORK
| 101 | 73 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK
| 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK
| 423 | 265 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 UP
| 209 | 139 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN
| 848 | 527 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN
| 1024 | 594 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN
| 637 | 377 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS
| 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS
| 734 | 436 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS
| 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS
| 775 | 494 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES
| 648 | 385 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 480 HELA
| 164 | 118 | All SZGR 2.0 genes in this pathway |
| SHIN B CELL LYMPHOMA CLUSTER 8
| 36 | 28 | All SZGR 2.0 genes in this pathway |
| MORI PRE BI LYMPHOCYTE UP
| 80 | 54 | All SZGR 2.0 genes in this pathway |
| MORI LARGE PRE BII LYMPHOCYTE UP
| 86 | 49 | All SZGR 2.0 genes in this pathway |
| MORI IMMATURE B LYMPHOCYTE DN
| 90 | 55 | All SZGR 2.0 genes in this pathway |
| COLLIS PRKDC SUBSTRATES
| 20 | 15 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES
| 230 | 137 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA DN
| 289 | 166 | All SZGR 2.0 genes in this pathway |
| LE EGR2 TARGETS UP
| 108 | 75 | All SZGR 2.0 genes in this pathway |
| REN BOUND BY E2F
| 61 | 40 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE UP
| 203 | 130 | All SZGR 2.0 genes in this pathway |
| ZHAN V2 LATE DIFFERENTIATION GENES
| 45 | 34 | All SZGR 2.0 genes in this pathway |
| GREENBAUM E2A TARGETS UP
| 33 | 18 | All SZGR 2.0 genes in this pathway |
| LY AGING PREMATURE DN
| 30 | 17 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR UP
| 240 | 152 | All SZGR 2.0 genes in this pathway |
| HU GENOTOXIC DAMAGE 24HR
| 35 | 22 | All SZGR 2.0 genes in this pathway |
| PAL PRMT5 TARGETS UP
| 203 | 135 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 8HR UP
| 105 | 73 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP
| 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| KIM GASTRIC CANCER CHEMOSENSITIVITY
| 103 | 64 | All SZGR 2.0 genes in this pathway |
| RHODES UNDIFFERENTIATED CANCER
| 69 | 44 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK UP
| 244 | 151 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS PEAK AT 24HR
| 43 | 30 | All SZGR 2.0 genes in this pathway |
| MODY HIPPOCAMPUS PRENATAL
| 42 | 33 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 4
| 48 | 32 | All SZGR 2.0 genes in this pathway |
| SONG TARGETS OF IE86 CMV PROTEIN
| 60 | 42 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION NEOCORTEX DN
| 88 | 58 | All SZGR 2.0 genes in this pathway |
| LY AGING OLD DN
| 56 | 35 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC UP
| 202 | 115 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 48HR DN
| 161 | 105 | All SZGR 2.0 genes in this pathway |
| LY AGING MIDDLE DN
| 16 | 10 | All SZGR 2.0 genes in this pathway |
| HU GENOTOXIC DAMAGE 4HR
| 35 | 28 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C3
| 20 | 15 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED
| 728 | 415 | All SZGR 2.0 genes in this pathway |
| AMUNDSON GAMMA RADIATION RESPONSE
| 40 | 25 | All SZGR 2.0 genes in this pathway |
| SARRIO EPITHELIAL MESENCHYMAL TRANSITION UP
| 180 | 114 | All SZGR 2.0 genes in this pathway |
| FUJII YBX1 TARGETS DN
| 202 | 132 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP
| 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP
| 863 | 514 | All SZGR 2.0 genes in this pathway |
| MATZUK SPERMATOCYTE
| 72 | 55 | All SZGR 2.0 genes in this pathway |
| WHITEFORD PEDIATRIC CANCER MARKERS
| 116 | 63 | All SZGR 2.0 genes in this pathway |
| CHEN HOXA5 TARGETS 9HR DN
| 41 | 17 | All SZGR 2.0 genes in this pathway |
| FRASOR RESPONSE TO SERM OR FULVESTRANT DN
| 50 | 29 | All SZGR 2.0 genes in this pathway |
| GOLDRATH ANTIGEN RESPONSE
| 346 | 192 | All SZGR 2.0 genes in this pathway |
| RUIZ TNC TARGETS DN
| 142 | 79 | All SZGR 2.0 genes in this pathway |
| CHANG CYCLING GENES
| 148 | 83 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN
| 435 | 289 | All SZGR 2.0 genes in this pathway |
| HOFFMANN LARGE TO SMALL PRE BII LYMPHOCYTE UP
| 163 | 102 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6
| 456 | 285 | All SZGR 2.0 genes in this pathway |
| CROONQUIST NRAS SIGNALING DN
| 72 | 47 | All SZGR 2.0 genes in this pathway |
| CROONQUIST NRAS VS STROMAL STIMULATION DN
| 99 | 65 | All SZGR 2.0 genes in this pathway |
| KOBAYASHI EGFR SIGNALING 24HR DN
| 251 | 151 | All SZGR 2.0 genes in this pathway |
| CHIARETTI T ALL RELAPSE PROGNOSIS
| 19 | 15 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC
| 1103 | 714 | All SZGR 2.0 genes in this pathway |
| DAZARD UV RESPONSE CLUSTER G5
| 12 | 8 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G2
| 182 | 102 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN
| 841 | 431 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 DN
| 374 | 217 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR UP
| 324 | 193 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE
| 1137 | 655 | All SZGR 2.0 genes in this pathway |
| LINSLEY MIR16 TARGETS
| 206 | 127 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP
| 570 | 339 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 7
| 403 | 240 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D
| 882 | 506 | All SZGR 2.0 genes in this pathway |
| ZHOU CELL CYCLE GENES IN IR RESPONSE 6HR
| 85 | 49 | All SZGR 2.0 genes in this pathway |
| ZHOU CELL CYCLE GENES IN IR RESPONSE 24HR
| 128 | 73 | All SZGR 2.0 genes in this pathway |
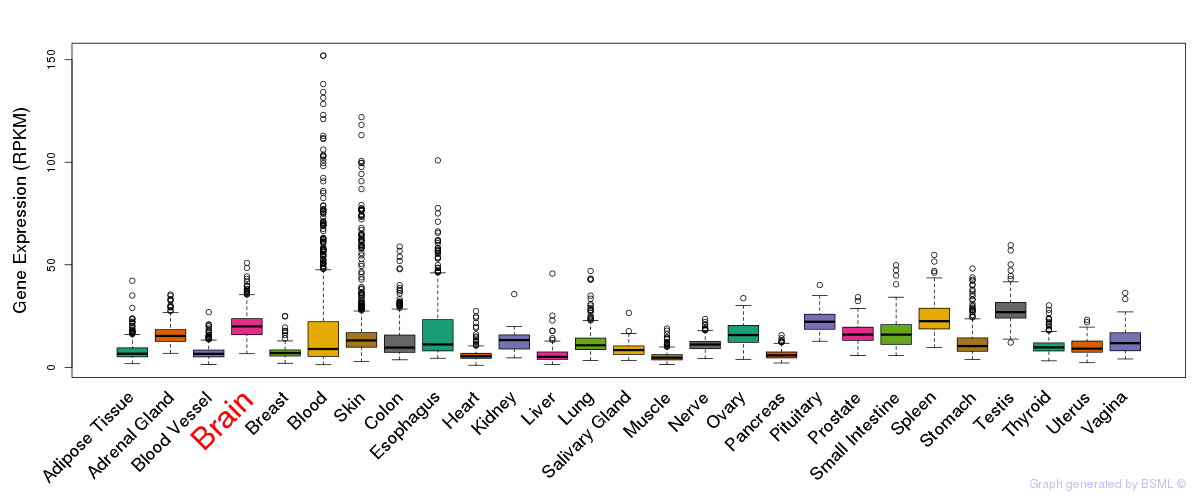
 Differentially methylated gene
Differentially methylated gene eQTL annotation
eQTL annotation