Gene Page: PGK1
Summary ?
| GeneID | 5230 |
| Symbol | PGK1 |
| Synonyms | HEL-S-68p|MIG10|PGKA |
| Description | phosphoglycerate kinase 1 |
| Reference | MIM:311800|HGNC:HGNC:8896|HPRD:02412| |
| Gene type | protein-coding |
| Map location | Xq13.3 |
| Sherlock p-value | 0.245 |
| Fetal beta | -1.309 |
| DMG | 1 (# studies) |
| Support | CELL METABOLISM G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 4 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Expression | Meta-analysis of gene expression | P value: 2.078 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg02148711 | X | 77041559 | PGK1 | 0.003 | -5.047 | DMG:vanEijk_2014 | |
| cg18449462 | X | 77155144 | PGK1 | 0.004 | -5.189 | DMG:vanEijk_2014 | |
| cg26848126 | X | 77582913 | PGK1 | 0.003 | -5.766 | DMG:vanEijk_2014 | |
| cg10526888 | X | 77151237 | PGK1 | 4.415E-4 | -5.784 | DMG:vanEijk_2014 |
Section II. Transcriptome annotation
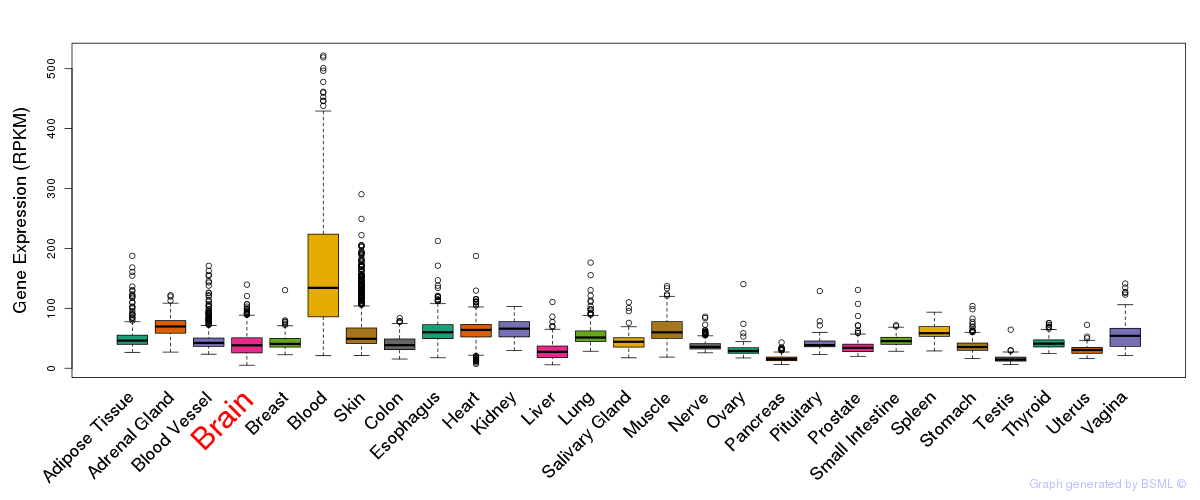
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0004618 | phosphoglycerate kinase activity | ISS | - | |
| GO:0005524 | ATP binding | ISS | - | |
| GO:0016740 | transferase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006096 | glycolysis | IEA | - | |
| GO:0016310 | phosphorylation | ISS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG GLYCOLYSIS GLUCONEOGENESIS | 62 | 41 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GLYCOLYSIS PATHWAY | 10 | 9 | All SZGR 2.0 genes in this pathway |
| PID HIF2PATHWAY | 34 | 29 | All SZGR 2.0 genes in this pathway |
| PID HIF1 TFPATHWAY | 66 | 52 | All SZGR 2.0 genes in this pathway |
| REACTOME GLYCOLYSIS | 29 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME GLUCONEOGENESIS | 34 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF CARBOHYDRATES | 247 | 154 | All SZGR 2.0 genes in this pathway |
| REACTOME GLUCOSE METABOLISM | 69 | 42 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA UP | 92 | 57 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| PUIFFE INVASION INHIBITED BY ASCITES UP | 82 | 51 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA UP | 305 | 185 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS DN | 310 | 188 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS DN | 384 | 230 | All SZGR 2.0 genes in this pathway |
| BOGNI TREATMENT RELATED MYELOID LEUKEMIA UP | 30 | 19 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY DN | 362 | 238 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 2HR UP | 27 | 19 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 6HR UP | 55 | 34 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA UP | 171 | 112 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA BY DMOG UP | 130 | 85 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HIF1A TARGETS DN | 91 | 58 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HIF1A AND HIF2A TARGETS DN | 104 | 72 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| KAN RESPONSE TO ARSENIC TRIOXIDE | 123 | 80 | All SZGR 2.0 genes in this pathway |
| DARWICHE SKIN TUMOR PROMOTER UP | 142 | 96 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK LOW UP | 162 | 104 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK HIGH UP | 147 | 101 | All SZGR 2.0 genes in this pathway |
| DARWICHE SQUAMOUS CELL CARCINOMA UP | 146 | 104 | All SZGR 2.0 genes in this pathway |
| HWANG PROSTATE CANCER MARKERS | 28 | 19 | All SZGR 2.0 genes in this pathway |
| APPIERTO RESPONSE TO FENRETINIDE DN | 51 | 35 | All SZGR 2.0 genes in this pathway |
| HUMMERICH SKIN CANCER PROGRESSION UP | 88 | 58 | All SZGR 2.0 genes in this pathway |
| LI LUNG CANCER | 41 | 30 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS LUMINAL | 326 | 213 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS NEUROEPITHELIUM UP | 176 | 111 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER UP | 227 | 137 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| GROSS HIF1A TARGETS DN | 25 | 15 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 AND HIF1A UP | 142 | 104 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 12HR UP | 116 | 79 | All SZGR 2.0 genes in this pathway |
| COLIN PILOCYTIC ASTROCYTOMA VS GLIOBLASTOMA DN | 28 | 21 | All SZGR 2.0 genes in this pathway |
| PENG LEUCINE DEPRIVATION DN | 187 | 122 | All SZGR 2.0 genes in this pathway |
| CHEN LUNG CANCER SURVIVAL | 28 | 22 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER METASTASIS DN | 121 | 65 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST DN | 309 | 206 | All SZGR 2.0 genes in this pathway |
| PENG GLUCOSE DEPRIVATION DN | 169 | 112 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE DN | 245 | 154 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| MENSE HYPOXIA UP | 98 | 71 | All SZGR 2.0 genes in this pathway |
| GREENBAUM E2A TARGETS UP | 33 | 18 | All SZGR 2.0 genes in this pathway |
| KAAB FAILED HEART ATRIUM DN | 141 | 99 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| HARRIS HYPOXIA | 81 | 64 | All SZGR 2.0 genes in this pathway |
| RAMASWAMY METASTASIS DN | 61 | 47 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN UP | 71 | 49 | All SZGR 2.0 genes in this pathway |
| LEONARD HYPOXIA | 47 | 35 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK UP | 244 | 151 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV SCC UP | 123 | 75 | All SZGR 2.0 genes in this pathway |
| DAZARD UV RESPONSE CLUSTER G1 | 67 | 41 | All SZGR 2.0 genes in this pathway |
| SEMENZA HIF1 TARGETS | 36 | 32 | All SZGR 2.0 genes in this pathway |
| JIANG AGING CEREBRAL CORTEX DN | 54 | 43 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| ZHONG SECRETOME OF LUNG CANCER AND MACROPHAGE | 77 | 50 | All SZGR 2.0 genes in this pathway |
| ZHONG SECRETOME OF LUNG CANCER AND FIBROBLAST | 132 | 93 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| SARRIO EPITHELIAL MESENCHYMAL TRANSITION UP | 180 | 114 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN UP | 181 | 112 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| LABBE WNT3A TARGETS DN | 97 | 53 | All SZGR 2.0 genes in this pathway |
| LABBE TARGETS OF TGFB1 AND WNT3A DN | 108 | 68 | All SZGR 2.0 genes in this pathway |
| NAKAMURA METASTASIS | 47 | 28 | All SZGR 2.0 genes in this pathway |
| NAKAMURA METASTASIS MODEL UP | 45 | 26 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA METAGENE | 242 | 168 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC | 420 | 269 | All SZGR 2.0 genes in this pathway |
| CHAUHAN RESPONSE TO METHOXYESTRADIOL DN | 102 | 65 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6 | 456 | 285 | All SZGR 2.0 genes in this pathway |
| SHAFFER IRF4 TARGETS IN MYELOMA VS MATURE B LYMPHOCYTE | 101 | 76 | All SZGR 2.0 genes in this pathway |
| SHAFFER IRF4 TARGETS IN ACTIVATED B LYMPHOCYTE | 81 | 66 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G3 UP | 188 | 121 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS UNANNOTATED DN | 193 | 112 | All SZGR 2.0 genes in this pathway |
| NGO MALIGNANT GLIOMA 1P LOH | 17 | 13 | All SZGR 2.0 genes in this pathway |
| FOURNIER ACINAR DEVELOPMENT LATE 2 | 277 | 172 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S1 | 237 | 159 | All SZGR 2.0 genes in this pathway |
| MOOTHA GLUCONEOGENESIS | 32 | 23 | All SZGR 2.0 genes in this pathway |
| MOOTHA GLYCOLYSIS | 21 | 13 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO GONADOTROPHINS UP | 91 | 59 | All SZGR 2.0 genes in this pathway |
| SASSON RESPONSE TO FORSKOLIN UP | 90 | 58 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| WANG ADIPOGENIC GENES REPRESSED BY SIRT1 | 28 | 21 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| VANDESLUIS COMMD1 TARGETS GROUP 3 UP | 89 | 50 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| BILANGES RAPAMYCIN SENSITIVE VIA TSC1 AND TSC2 | 73 | 37 | All SZGR 2.0 genes in this pathway |
| FARDIN HYPOXIA 11 | 32 | 29 | All SZGR 2.0 genes in this pathway |
| SERVITJA ISLET HNF1A TARGETS DN | 109 | 71 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| KUMAR PATHOGEN LOAD BY MACROPHAGES | 275 | 155 | All SZGR 2.0 genes in this pathway |
| KUMAR AUTOPHAGY NETWORK | 71 | 46 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-143 | 151 | 157 | 1A | hsa-miR-143brain | UGAGAUGAAGCACUGUAGCUCA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.