Gene Page: RBL1
Summary ?
| GeneID | 5933 |
| Symbol | RBL1 |
| Synonyms | CP107|PRB1|p107 |
| Description | retinoblastoma-like 1 |
| Reference | MIM:116957|HGNC:HGNC:9893|Ensembl:ENSG00000080839|HPRD:00312|Vega:OTTHUMG00000032406 |
| Gene type | protein-coding |
| Map location | 20q11.2 |
| Pascal p-value | 0.222 |
| Fetal beta | 1.471 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg21570468 | 20 | 35724588 | RBL1 | 5.41E-8 | -0.005 | 1.4E-5 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
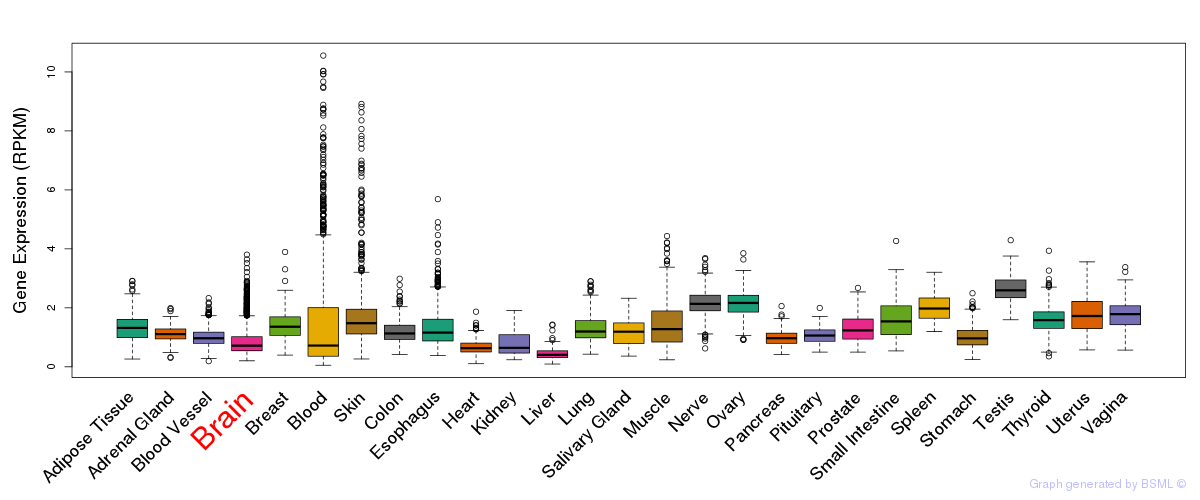
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KIAA1429 | 0.93 | 0.93 |
| CEBPZ | 0.93 | 0.93 |
| PRPF40A | 0.92 | 0.93 |
| XPO1 | 0.92 | 0.93 |
| RMI1 | 0.92 | 0.91 |
| N4BP2L2 | 0.92 | 0.92 |
| DHX15 | 0.92 | 0.92 |
| NUP54 | 0.92 | 0.92 |
| CRNKL1 | 0.92 | 0.91 |
| USP1 | 0.91 | 0.92 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.69 | -0.83 |
| AF347015.31 | -0.69 | -0.83 |
| AF347015.33 | -0.67 | -0.80 |
| AF347015.27 | -0.67 | -0.80 |
| MT-CYB | -0.67 | -0.81 |
| AF347015.8 | -0.67 | -0.82 |
| AF347015.15 | -0.66 | -0.82 |
| PTH1R | -0.66 | -0.76 |
| HLA-F | -0.65 | -0.74 |
| IFI27 | -0.65 | -0.82 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AATF | CHE-1 | CHE1 | DED | apoptosis antagonizing transcription factor | Affinity Capture-Western | BioGRID | 12450794 |
| ARID4A | RBBP1 | RBP-1 | RBP1 | AT rich interactive domain 4A (RBP1-like) | Affinity Capture-Western | BioGRID | 10490602 |
| BEGAIN | KIAA1446 | brain-enriched guanylate kinase-associated homolog (rat) | Affinity Capture-Western Two-hybrid | BioGRID | 16189514 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | Co-localization Reconstituted Complex | BioGRID | 11521194 |
| BRF1 | BRF | FLJ42674 | FLJ43034 | GTF3B | MGC105048 | TAF3B2 | TAF3C | TAFIII90 | TF3B90 | TFIIIB90 | hBRF | BRF1 homolog, subunit of RNA polymerase III transcription initiation factor IIIB (S. cerevisiae) | - | HPRD,BioGRID | 10330166 |
| CCNA1 | - | cyclin A1 | Reconstituted Complex | BioGRID | 10022926 |
| CCNA2 | CCN1 | CCNA | cyclin A2 | - | HPRD,BioGRID | 1532458 |8230483 |12439743 |
| CCND1 | BCL1 | D11S287E | PRAD1 | U21B31 | cyclin D1 | - | HPRD,BioGRID | 8643455 |
| CCNE1 | CCNE | cyclin E1 | - | HPRD | 8230483 |
| CCNE1 | CCNE | cyclin E1 | Affinity Capture-Western | BioGRID | 9891079 |
| CDK2 | p33(CDK2) | cyclin-dependent kinase 2 | Biochemical Activity in vitro in vivo | BioGRID | 9891079 |11884610 |
| CDK2 | p33(CDK2) | cyclin-dependent kinase 2 | - | HPRD | 8230483|11884610 |
| CDKN1C | BWCR | BWS | KIP2 | WBS | p57 | cyclin-dependent kinase inhibitor 1C (p57, Kip2) | Affinity Capture-Western | BioGRID | 12947099 |
| CREG1 | CREG | cellular repressor of E1A-stimulated genes 1 | - | HPRD,BioGRID | 9710587 |
| E2F1 | E2F-1 | RBAP1 | RBBP3 | RBP3 | E2F transcription factor 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 8230483 |
| E2F1 | E2F-1 | RBAP1 | RBBP3 | RBP3 | E2F transcription factor 1 | E2F1 interacts with the p107 promoter region. | BIND | 10766737 |
| E2F2 | E2F-2 | E2F transcription factor 2 | E2F2 interacts with the p107 promoter region. | BIND | 10766737 |
| E2F4 | E2F-4 | E2F transcription factor 4, p107/p130-binding | E2F4 interacts with the p107 promoter region. | BIND | 10766737 |
| E2F4 | E2F-4 | E2F transcription factor 4, p107/p130-binding | p107 interacts with E2F4. | BIND | 8816797 |
| E2F4 | E2F-4 | E2F transcription factor 4, p107/p130-binding | - | HPRD,BioGRID | 8643455 |11573202 |12150994 |
| E2F5 | E2F-5 | E2F transcription factor 5, p130-binding | Two-hybrid | BioGRID | 7760804 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | - | HPRD,BioGRID | 9724731 |
| HDAC2 | RPD3 | YAF1 | histone deacetylase 2 | Affinity Capture-Western | BioGRID | 10490602 |
| HDAC3 | HD3 | RPD3 | RPD3-2 | histone deacetylase 3 | Affinity Capture-Western | BioGRID | 10490602 |
| JARID1A | KDM5A | RBBP2 | RBP2 | jumonji, AT rich interactive domain 1A | - | HPRD,BioGRID | 7935440 |
| LIN9 | BARA | BARPsv | Lin-9 | TGS | TGS1 | lin-9 homolog (C. elegans) | hMip130/TWIT interacts with p107. | BIND | 15479636 |
| MCM7 | CDABP0042 | CDC47 | MCM2 | P1.1-MCM3 | P1CDC47 | P85MCM | PNAS-146 | minichromosome maintenance complex component 7 | - | HPRD,BioGRID | 9566894 |
| MCM7 | CDABP0042 | CDC47 | MCM2 | P1.1-MCM3 | P1CDC47 | P85MCM | PNAS-146 | minichromosome maintenance complex component 7 | MCM7 interacts with p107. | BIND | 9566894 |
| MT1G | MGC12386 | MT1 | MT1K | metallothionein 1G | RBL1 (P107) interacts with the MT1G promoter. | BIND | 15735762 |
| MYBL2 | B-MYB | BMYB | MGC15600 | v-myb myeloblastosis viral oncogene homolog (avian)-like 2 | - | HPRD,BioGRID | 12439743 |12947099 |
| MYC | bHLHe39 | c-Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | - | HPRD,BioGRID | 8076603 |
| PHB | PHB1 | prohibitin | in vitro in vivo | BioGRID | 10376528 |
| PPP2R3A | PPP2R3 | PR130 | PR72 | protein phosphatase 2 (formerly 2A), regulatory subunit B'', alpha | Affinity Capture-Western | BioGRID | 9927208 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | Rb interacts with the p107 promoter. | BIND | 15716956 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | Affinity Capture-Western | BioGRID | 10490602 |
| RBBP8 | CTIP | RIM | retinoblastoma binding protein 8 | - | HPRD,BioGRID | 9721205 |
| RBBP9 | BOG | MGC9236 | RBBP10 | retinoblastoma binding protein 9 | - | HPRD,BioGRID | 9697699 |
| RBL2 | FLJ26459 | P130 | Rb2 | retinoblastoma-like 2 (p130) | p130 interacts with the p107 promoter region. | BIND | 10766737 |
| SMAD2 | JV18 | JV18-1 | MADH2 | MADR2 | MGC22139 | MGC34440 | hMAD-2 | hSMAD2 | SMAD family member 2 | - | HPRD | 12150994 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | - | HPRD,BioGRID | 12150994 |
| SMAD4 | DPC4 | JIP | MADH4 | SMAD family member 4 | - | HPRD,BioGRID | 12150994 |
| SNW1 | Bx42 | MGC119379 | NCOA-62 | PRPF45 | Prp45 | SKIIP | SKIP | SNW domain containing 1 | - | HPRD,BioGRID | 12466551 |
| SUV39H1 | KMT1A | MG44 | SUV39H | suppressor of variegation 3-9 homolog 1 (Drosophila) | Affinity Capture-Western | BioGRID | 12588981 |
| TAF1 | BA2R | CCG1 | CCGS | DYT3 | KAT4 | N-TAF1 | NSCL2 | OF | P250 | TAF2A | TAFII250 | TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa | - | HPRD,BioGRID | 9242374 |
| TFDP1 | DP1 | DRTF1 | Dp-1 | transcription factor Dp-1 | Affinity Capture-Western | BioGRID | 7739537 |
| TFDP2 | DP2 | Dp-2 | transcription factor Dp-2 (E2F dimerization partner 2) | Affinity Capture-Western | BioGRID | 7739537 |
| USP4 | MGC149848 | MGC149849 | UNP | Unph | ubiquitin specific peptidase 4 (proto-oncogene) | in vitro in vivo | BioGRID | 11571651 |
| UXT | ART-27 | ubiquitously-expressed transcript | p107 interacts with the UXT chromatin. | BIND | 11799066 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CELL CYCLE | 128 | 84 | All SZGR 2.0 genes in this pathway |
| KEGG TGF BETA SIGNALING PATHWAY | 86 | 64 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CELLCYCLE PATHWAY | 23 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ETS PATHWAY | 18 | 12 | All SZGR 2.0 genes in this pathway |
| PID SMAD2 3NUCLEAR PATHWAY | 82 | 63 | All SZGR 2.0 genes in this pathway |
| PID E2F PATHWAY | 74 | 48 | All SZGR 2.0 genes in this pathway |
| PID MYC REPRESS PATHWAY | 63 | 52 | All SZGR 2.0 genes in this pathway |
| REACTOME G0 AND EARLY G1 | 25 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | 27 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | 38 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME GENERIC TRANSCRIPTION PATHWAY | 352 | 181 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME G1 PHASE | 38 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC G1 G1 S PHASES | 137 | 79 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY TGF BETA RECEPTOR COMPLEX | 63 | 42 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED UP | 633 | 376 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION KRAS UP | 126 | 72 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 DN | 378 | 231 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 UP | 329 | 196 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 CHRONIC LOF UP | 115 | 78 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF DN | 228 | 137 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| STREICHER LSM1 TARGETS UP | 44 | 34 | All SZGR 2.0 genes in this pathway |
| STREICHER LSM1 TARGETS DN | 19 | 13 | All SZGR 2.0 genes in this pathway |
| KONG E2F3 TARGETS | 97 | 58 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION | 77 | 51 | All SZGR 2.0 genes in this pathway |
| HERNANDEZ MITOTIC ARREST BY DOCETAXEL 1 DN | 38 | 23 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS NEUROEPITHELIUM DN | 164 | 111 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION UP | 69 | 55 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES CORE NINE CORRELATED | 100 | 68 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO IKK INHIBITOR AND TNF DN | 103 | 64 | All SZGR 2.0 genes in this pathway |
| MORI SMALL PRE BII LYMPHOCYTE DN | 76 | 52 | All SZGR 2.0 genes in this pathway |
| LE EGR2 TARGETS UP | 108 | 75 | All SZGR 2.0 genes in this pathway |
| REN BOUND BY E2F | 61 | 40 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA CD1 AND CD2 UP | 89 | 51 | All SZGR 2.0 genes in this pathway |
| KAMMINGA EZH2 TARGETS | 41 | 26 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP | 390 | 242 | All SZGR 2.0 genes in this pathway |
| PAL PRMT5 TARGETS UP | 203 | 135 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE UP | 244 | 165 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| CHEN ETV5 TARGETS TESTIS | 23 | 14 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE DN | 373 | 196 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN | 409 | 268 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY GAMMA AND UV RADIATION | 88 | 65 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE DN | 195 | 135 | All SZGR 2.0 genes in this pathway |
| SARRIO EPITHELIAL MESENCHYMAL TRANSITION UP | 180 | 114 | All SZGR 2.0 genes in this pathway |
| DE YY1 TARGETS DN | 92 | 64 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS UP | 425 | 253 | All SZGR 2.0 genes in this pathway |
| ZHENG GLIOBLASTOMA PLASTICITY UP | 250 | 168 | All SZGR 2.0 genes in this pathway |
| HOFFMANN LARGE TO SMALL PRE BII LYMPHOCYTE UP | 163 | 102 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6 | 456 | 285 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR DN | 185 | 116 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR DN | 277 | 166 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS DN | 89 | 50 | All SZGR 2.0 genes in this pathway |
| ONO AML1 TARGETS DN | 41 | 25 | All SZGR 2.0 genes in this pathway |
| ONO FOXP3 TARGETS DN | 42 | 23 | All SZGR 2.0 genes in this pathway |
| PYEON CANCER HEAD AND NECK VS CERVICAL UP | 193 | 95 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 6HR UP | 229 | 149 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR UP | 324 | 193 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| YU BAP1 TARGETS | 29 | 21 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM A | 182 | 108 | All SZGR 2.0 genes in this pathway |