Gene Page: SDC2
Summary ?
| GeneID | 6383 |
| Symbol | SDC2 |
| Synonyms | CD362|HSPG|HSPG1|SYND2 |
| Description | syndecan 2 |
| Reference | MIM:142460|HGNC:HGNC:10659|Ensembl:ENSG00000169439|HPRD:00803|Vega:OTTHUMG00000164689 |
| Gene type | protein-coding |
| Map location | 8q22-q23 |
| Pascal p-value | 0.038 |
| Fetal beta | -1.198 |
| DMG | 2 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 2 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0329 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg14538332 | 8 | 97506180 | SDC2 | -0.025 | 0.37 | DMG:Nishioka_2013 | |
| cg16673702 | 8 | 97507592 | SDC2 | 7.79E-8 | -0.01 | 1.84E-5 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs9882137 | chr3 | 29494146 | SDC2 | 6383 | 0.01 | trans | ||
| rs9882501 | chr3 | 29494429 | SDC2 | 6383 | 0.07 | trans | ||
| snp_a-1789759 | 0 | SDC2 | 6383 | 0.14 | trans | |||
| rs17139868 | chr7 | 117079228 | SDC2 | 6383 | 0.01 | trans |
Section II. Transcriptome annotation
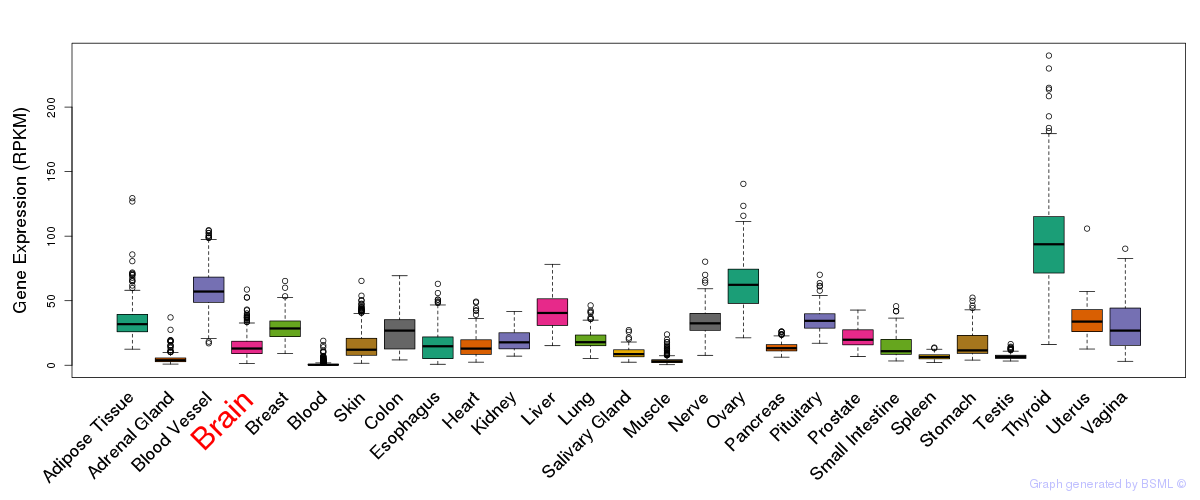
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ZNF416 | 0.92 | 0.88 |
| PRPSAP2 | 0.92 | 0.89 |
| WDR70 | 0.92 | 0.90 |
| BZW2 | 0.91 | 0.92 |
| ZNF555 | 0.91 | 0.85 |
| RFWD2 | 0.91 | 0.90 |
| YTHDF2 | 0.91 | 0.84 |
| C12orf41 | 0.91 | 0.90 |
| EIF4A1 | 0.90 | 0.90 |
| ZNF259 | 0.90 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.33 | -0.73 | -0.87 |
| AF347015.27 | -0.72 | -0.84 |
| MT-CO2 | -0.72 | -0.83 |
| MT-CYB | -0.72 | -0.86 |
| AF347015.31 | -0.72 | -0.83 |
| AIFM3 | -0.71 | -0.79 |
| AF347015.8 | -0.70 | -0.85 |
| HLA-F | -0.70 | -0.76 |
| HEPN1 | -0.70 | -0.77 |
| AF347015.15 | -0.70 | -0.85 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003674 | molecular_function | ND | - | |
| GO:0008092 | cytoskeletal protein binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008150 | biological_process | ND | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | NAS | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| APOE | AD2 | LPG | MGC1571 | apoprotein | apolipoprotein E | - | HPRD | 9869645 |
| CASK | CAGH39 | CMG | FLJ22219 | FLJ31914 | LIN2 | MICPCH | TNRC8 | calcium/calmodulin-dependent serine protein kinase (MAGUK family) | - | HPRD,BioGRID | 9660868 |
| CSF2 | GMCSF | MGC131935 | MGC138897 | colony stimulating factor 2 (granulocyte-macrophage) | - | HPRD,BioGRID | 10734053 |
| CSF2RA | CD116 | CDw116 | CSF2R | CSF2RAX | CSF2RAY | CSF2RX | CSF2RY | GM-CSF-R-alpha | GMCSFR | GMR | MGC3848 | MGC4838 | colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) | Affinity Capture-Western | BioGRID | 10734053 |
| EPB41L1 | 4.1N | DKFZp686H17242 | KIAA0338 | MGC11072 | erythrocyte membrane protein band 4.1-like 1 | Affinity Capture-Western | BioGRID | 12676536 |
| EZR | CVIL | CVL | DKFZp762H157 | FLJ26216 | MGC1584 | VIL2 | ezrin | - | HPRD,BioGRID | 10704377 |
| FGF2 | BFGF | FGFB | HBGF-2 | fibroblast growth factor 2 (basic) | Co-purification | BioGRID | 16982797 |
| FN1 | CIG | DKFZp686F10164 | DKFZp686H0342 | DKFZp686I1370 | DKFZp686O13149 | ED-B | FINC | FN | FNZ | GFND | GFND2 | LETS | MSF | fibronectin 1 | - | HPRD,BioGRID | 10772816 |
| HGF | F-TCF | HGFB | HPTA | SF | hepatocyte growth factor (hepapoietin A; scatter factor) | - | HPRD,BioGRID | 8157651 |
| ITPR1 | INSP3R1 | IP3R | IP3R1 | SCA15 | SCA16 | inositol 1,4,5-triphosphate receptor, type 1 | Affinity Capture-Western | BioGRID | 12676536 |
| KAL1 | ADMLX | HHA | KAL | KALIG-1 | KMS | Kallmann syndrome 1 sequence | - | HPRD,BioGRID | 8832397 |
| LAMA3 | BM600 | E170 | LAMNA | LOCS | lama3a | laminin, alpha 3 | - | HPRD,BioGRID | 11373281 |
| PF4 | CXCL4 | MGC138298 | SCYB4 | platelet factor 4 | - | HPRD,BioGRID | 7524669 |
| REG3A | HIP | INGAP | PAP | PAP-H | PAP1 | PBCGF | REG-III | REG3 | regenerating islet-derived 3 alpha | - | HPRD,BioGRID | 8997243 |
| SDCBP | MDA-9 | ST1 | SYCL | TACIP18 | syndecan binding protein (syntenin) | - | HPRD,BioGRID | 9391086 |
| SERPINC1 | AT3 | ATIII | MGC22579 | serpin peptidase inhibitor, clade C (antithrombin), member 1 | - | HPRD | 1315572 |
| SPARC | ON | secreted protein, acidic, cysteine-rich (osteonectin) | - | HPRD | 2745554 |
| TRAPPC4 | CGI-104 | HSPC172 | PTD009 | SBDN | SYNBINDIN | TRS23 | trafficking protein particle complex 4 | - | HPRD | 11018053 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ECM RECEPTOR INTERACTION | 84 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG CELL ADHESION MOLECULES CAMS | 134 | 93 | All SZGR 2.0 genes in this pathway |
| PID SHP2 PATHWAY | 58 | 46 | All SZGR 2.0 genes in this pathway |
| PID SYNDECAN 2 PATHWAY | 33 | 27 | All SZGR 2.0 genes in this pathway |
| PID FGF PATHWAY | 55 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME HS GAG DEGRADATION | 20 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | 49 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME HS GAG BIOSYNTHESIS | 31 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME HEPARAN SULFATE HEPARIN HS GAG METABOLISM | 52 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME GLYCOSAMINOGLYCAN METABOLISM | 111 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | 25 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF CARBOHYDRATES | 247 | 154 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE UP | 351 | 230 | All SZGR 2.0 genes in this pathway |
| DAVICIONI TARGETS OF PAX FOXO1 FUSIONS UP | 255 | 177 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP | 579 | 346 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION MONOCYTE DN | 54 | 38 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS DN | 232 | 139 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC2 TARGETS DN | 133 | 77 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN | 536 | 332 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP DN | 199 | 124 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER DN | 232 | 154 | All SZGR 2.0 genes in this pathway |
| HERNANDEZ MITOTIC ARREST BY DOCETAXEL 1 DN | 38 | 23 | All SZGR 2.0 genes in this pathway |
| SASAI RESISTANCE TO NEOPLASTIC TRANSFROMATION | 50 | 31 | All SZGR 2.0 genes in this pathway |
| HASINA NOL7 TARGETS DN | 13 | 9 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| BALDWIN PRKCI TARGETS UP | 35 | 26 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| WANG PROSTATE CANCER ANDROGEN INDEPENDENT | 66 | 37 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 8Q12 Q22 AMPLICON | 132 | 82 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 UP | 256 | 159 | All SZGR 2.0 genes in this pathway |
| JECHLINGER EPITHELIAL TO MESENCHYMAL TRANSITION UP | 71 | 51 | All SZGR 2.0 genes in this pathway |
| FRASOR RESPONSE TO ESTRADIOL UP | 37 | 28 | All SZGR 2.0 genes in this pathway |
| SMITH LIVER CANCER | 45 | 27 | All SZGR 2.0 genes in this pathway |
| LENAOUR DENDRITIC CELL MATURATION DN | 128 | 90 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA CD2 UP | 45 | 32 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| DELASERNA MYOD TARGETS DN | 57 | 42 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN UP | 262 | 186 | All SZGR 2.0 genes in this pathway |
| TSENG IRS1 TARGETS UP | 113 | 71 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS UP | 280 | 183 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 12HR DN | 101 | 64 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| CLASPER LYMPHATIC VESSELS DURING METASTASIS DN | 36 | 23 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL UP | 260 | 174 | All SZGR 2.0 genes in this pathway |
| RAY TUMORIGENESIS BY ERBB2 CDC25A DN | 159 | 105 | All SZGR 2.0 genes in this pathway |
| GU PDEF TARGETS UP | 71 | 49 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP | 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 4HR DN | 254 | 158 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| WINZEN DEGRADED VIA KHSRP | 100 | 70 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 7 | 403 | 240 | All SZGR 2.0 genes in this pathway |
| NABA ECM AFFILIATED | 171 | 89 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124.1 | 1928 | 1934 | 1A | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-182 | 2192 | 2198 | 1A | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-186 | 157 | 163 | 1A | hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU |
| miR-200bc/429 | 757 | 763 | m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-218 | 988 | 994 | m8 | hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU |
| miR-25/32/92/363/367 | 465 | 471 | m8 | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| miR-27 | 201 | 207 | 1A | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-376c | 917 | 924 | 1A,m8 | hsa-miR-376c | AACAUAGAGGAAAUUCCACG |
| miR-485-5p | 731 | 737 | 1A | hsa-miR-485-5p | AGAGGCUGGCCGUGAUGAAUUC |
| miR-493-5p | 1131 | 1137 | m8 | hsa-miR-493-5p | UUGUACAUGGUAGGCUUUCAUU |
| miR-499 | 1019 | 1025 | m8 | hsa-miR-499 | UUAAGACUUGCAGUGAUGUUUAA |
| miR-500 | 464 | 470 | 1A | hsa-miR-500 | AUGCACCUGGGCAAGGAUUCUG |
| miR-9 | 1428 | 1434 | 1A | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
| hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA | ||||
| miR-96 | 2191 | 2198 | 1A,m8 | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.