Gene Page: TCF3
Summary ?
| GeneID | 6929 |
| Symbol | TCF3 |
| Synonyms | E2A|E47|ITF1|TCF-3|VDIR|bHLHb21 |
| Description | transcription factor 3 |
| Reference | MIM:147141|HGNC:HGNC:11633|Ensembl:ENSG00000071564|HPRD:00918|Vega:OTTHUMG00000180031 |
| Gene type | protein-coding |
| Map location | 19p13.3 |
| Pascal p-value | 0.012 |
| Fetal beta | 0.472 |
| DMG | 2 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Montano_2016 | Genome-wide DNA methylation analysis | This dataset includes 172 replicated associations between CpGs with schizophrenia. | 2 |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg23009327 | 19 | 1630248 | TCF3 | 1.32E-6 | 0.01 | 0.017 | DMG:Montano_2016 |
| cg21030400 | 19 | 2052564 | TCF3 | 2.31E-4 | -3.744 | DMG:vanEijk_2014 |
Section II. Transcriptome annotation
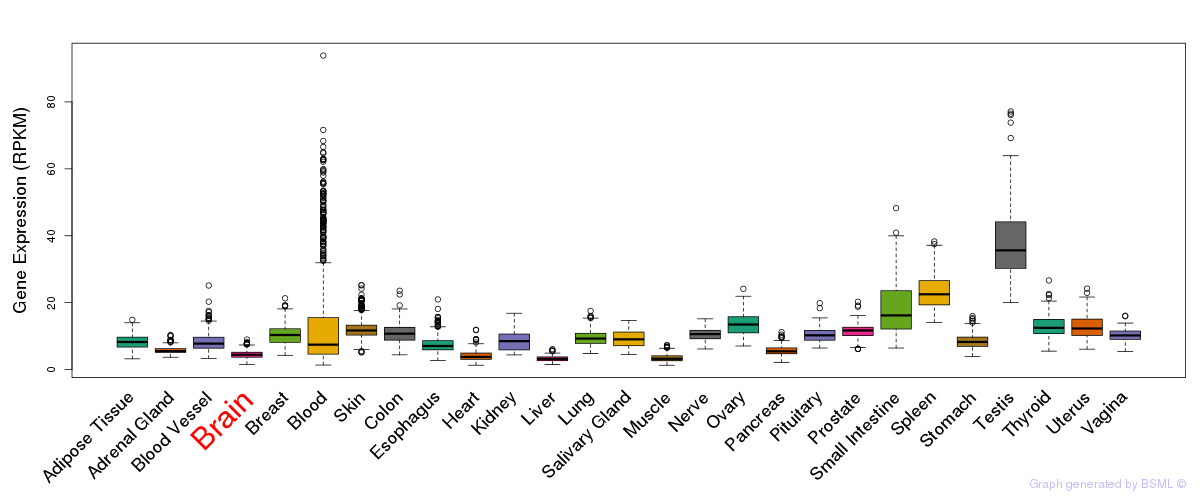
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ASCL3 | HASH3 | SGN1 | bHLHa42 | achaete-scute complex homolog 3 (Drosophila) | - | HPRD | 11784080 |
| BHLHB8 | MIST1 | bHLHa15 | basic helix-loop-helix domain containing, class B, 8 | - | HPRD | 9073453 |
| CALM1 | CALML2 | CAMI | DD132 | PHKD | calmodulin 1 (phosphorylase kinase, delta) | - | HPRD,BioGRID | 10757985 |
| CALM3 | PHKD | PHKD3 | calmodulin 3 (phosphorylase kinase, delta) | - | HPRD | 10757985 |
| CBFA2T3 | ETO2 | MTG16 | MTGR2 | ZMYND4 | core-binding factor, runt domain, alpha subunit 2; translocated to, 3 | Affinity Capture-Western Reconstituted Complex | BioGRID | 16407974 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | - | HPRD,BioGRID | 12435739 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | - | HPRD | 12107263 |
| ELK3 | ERP | NET | SAP2 | ELK3, ETS-domain protein (SRF accessory protein 2) | - | HPRD,BioGRID | 8918463 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | - | HPRD,BioGRID | 12435739 |
| FERD3L | MGC119861 | N-TWIST | NATO3 | NTWIST | Fer3-like (Drosophila) | - | HPRD | 12217327 |
| GARNL1 | DKFZp566D133 | DKFZp667F074 | GRIPE | KIAA0884 | TULIP1 | GTPase activating Rap/RanGAP domain-like 1 | - | HPRD,BioGRID | 12200424 |
| HAND1 | Hxt | Thing1 | bHLHa27 | eHand | heart and neural crest derivatives expressed 1 | - | HPRD | 10924525 |11802795 |
| HAND2 | DHAND2 | FLJ16260 | Hed | MGC125303 | MGC125304 | Thing2 | bHLHa26 | dHand | heart and neural crest derivatives expressed 2 | - | HPRD,BioGRID | 11812799 |
| ID1 | ID | bHLHb24 | inhibitor of DNA binding 1, dominant negative helix-loop-helix protein | Two-hybrid | BioGRID | 9242638 |
| ID2 | GIG8 | ID2A | ID2H | MGC26389 | bHLHb26 | inhibitor of DNA binding 2, dominant negative helix-loop-helix protein | - | HPRD,BioGRID | 9242638 |
| ID3 | HEIR-1 | bHLHb25 | inhibitor of DNA binding 3, dominant negative helix-loop-helix protein | - | HPRD | 8759016 |9525934 |
| ID3 | HEIR-1 | bHLHb25 | inhibitor of DNA binding 3, dominant negative helix-loop-helix protein | Reconstituted Complex Two-hybrid | BioGRID | 9242638 |9525934 |
| KAT2B | CAF | P | P/CAF | PCAF | K(lysine) acetyltransferase 2B | - | HPRD,BioGRID | 12435739 |
| LDB1 | CLIM2 | NLI | LIM domain binding 1 | Affinity Capture-Western | BioGRID | 16407974 |
| LMX1A | LMX-1 | LMX1 | LMX1.1 | MGC87616 | LIM homeobox transcription factor 1, alpha | Reconstituted Complex Two-hybrid | BioGRID | 9199284 |
| LYL1 | bHLHa18 | lymphoblastic leukemia derived sequence 1 | - | HPRD,BioGRID | 8628307 |
| MAPKAPK2 | MK2 | mitogen-activated protein kinase-activated protein kinase 2 | - | HPRD,BioGRID | 10781029 |
| MAPKAPK3 | 3PK | MAPKAP3 | mitogen-activated protein kinase-activated protein kinase 3 | - | HPRD,BioGRID | 10781029 |
| MSC | ABF-1 | ABF1 | MYOR | bHLHa22 | musculin (activated B-cell factor-1) | - | HPRD | 9584154 |
| MYF5 | bHLHc2 | myogenic factor 5 | Two-hybrid | BioGRID | 9242638 |
| MYF6 | MRF4 | bHLHc4 | myogenic factor 6 (herculin) | Two-hybrid | BioGRID | 9242638 |
| MYOD1 | MYF3 | MYOD | PUM | bHLHc1 | myogenic differentiation 1 | - | HPRD | 1649701 |9001211 |9184158 |
| MYOD1 | MYF3 | MYOD | PUM | bHLHc1 | myogenic differentiation 1 | in vitro in vivo Reconstituted Complex Two-hybrid | BioGRID | 1649701 |9184158 |9242638 |
| MYOG | MYF4 | MYOGENIN | bHLHc3 | myogenin (myogenic factor 4) | in vitro in vivo Two-hybrid | BioGRID | 1325437 |2163343 |9242638 |
| MYOG | MYF4 | MYOGENIN | bHLHc3 | myogenin (myogenic factor 4) | - | HPRD | 1649701 |
| NEDD9 | CAS-L | CAS2 | CASL | CASS2 | HEF1 | dJ49G10.2 | dJ761I2.1 | neural precursor cell expressed, developmentally down-regulated 9 | - | HPRD | 10502414 |
| NHLH1 | HEN1 | NSCL | NSCL1 | bHLHa35 | nescient helix loop helix 1 | Two-hybrid | BioGRID | 12878195 |
| PDX1 | GSF | IDX-1 | IPF1 | IUF1 | MODY4 | PDX-1 | STF-1 | pancreatic and duodenal homeobox 1 | - | HPRD,BioGRID | 10636926 |
| PSMD4 | AF | AF-1 | ASF | MCB1 | Rpn10 | S5A | pUB-R5 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 4 | - | HPRD,BioGRID | 9235903 |
| PSMD9 | MGC8644 | Rpn4 | p27 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 | - | HPRD | 10567574 |
| PTF1A | PTF1-p48 | bHLHa29 | pancreas specific transcription factor, 1a | - | HPRD,BioGRID | 1720355 |10768861 |
| RUNX1T1 | AML1T1 | CBFA2T1 | CDR | ETO | MGC2796 | MTG8 | MTG8b | ZMYND2 | runt-related transcription factor 1; translocated to, 1 (cyclin D-related) | ETO interacts with E2A. | BIND | 15333839 |
| SKP2 | FBL1 | FBXL1 | FLB1 | MGC1366 | S-phase kinase-associated protein 2 (p45) | - | HPRD,BioGRID | 14592976 |
| TAL1 | SCL | TCL5 | bHLHa17 | tal-1 | T-cell acute lymphocytic leukemia 1 | - | HPRD | 8159721|9507011 |
| TAL1 | SCL | TCL5 | bHLHa17 | tal-1 | T-cell acute lymphocytic leukemia 1 | Affinity Capture-MS Affinity Capture-Western in vitro in vivo Reconstituted Complex Two-hybrid | BioGRID | 8159721 |16407974 |
| TAL2 | bHLHa19 | T-cell acute lymphocytic leukemia 2 | - | HPRD | 8152805 |
| TCF12 | HEB | HTF4 | HsT17266 | bHLHb20 | transcription factor 12 | - | HPRD,BioGRID | 1446075 |12566462 |
| TCF3 | E2A | ITF1 | MGC129647 | MGC129648 | bHLHb21 | transcription factor 3 (E2A immunoglobulin enhancer binding factors E12/E47) | Co-fractionation | BioGRID | 12435739 |
| TCF3 | E2A | ITF1 | MGC129647 | MGC129648 | bHLHb21 | transcription factor 3 (E2A immunoglobulin enhancer binding factors E12/E47) | - | HPRD | 2503252 |7623842 |
| TCF4 | E2-2 | ITF2 | MGC149723 | MGC149724 | PTHS | SEF2 | SEF2-1 | SEF2-1A | SEF2-1B | bHLHb19 | transcription factor 4 | - | HPRD | 8999959 |
| TFPT | FB1 | INO80F | amida | TCF3 (E2A) fusion partner (in childhood Leukemia) | - | HPRD | 10086727 |
| TWIST1 | ACS3 | BPES2 | BPES3 | SCS | TWIST | bHLHa38 | twist homolog 1 (Drosophila) | - | HPRD,BioGRID | 10749989 |
| UBE2I | C358B7.1 | P18 | UBC9 | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | Reconstituted Complex Two-hybrid | BioGRID | 10497239 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID SMAD2 3NUCLEAR PATHWAY | 82 | 63 | All SZGR 2.0 genes in this pathway |
| PID P38 MK2 PATHWAY | 21 | 19 | All SZGR 2.0 genes in this pathway |
| PID HES HEY PATHWAY | 48 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME MYOGENESIS | 28 | 20 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP | 579 | 346 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS UP | 372 | 227 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED DN | 335 | 193 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 DN | 281 | 186 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN | 536 | 332 | All SZGR 2.0 genes in this pathway |
| HUMMEL BURKITTS LYMPHOMA UP | 43 | 27 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| PROVENZANI METASTASIS UP | 194 | 112 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| KLEIN TARGETS OF BCR ABL1 FUSION | 45 | 34 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM5 | 94 | 59 | All SZGR 2.0 genes in this pathway |
| PUJANA XPRSS INT NETWORK | 168 | 103 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA CENTERED NETWORK | 117 | 72 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS | 539 | 324 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| MORI EMU MYC LYMPHOMA BY ONSET TIME UP | 110 | 69 | All SZGR 2.0 genes in this pathway |
| GOLUB ALL VS AML UP | 24 | 20 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| HADDAD B LYMPHOCYTE PROGENITOR | 293 | 193 | All SZGR 2.0 genes in this pathway |
| BASSO CD40 SIGNALING DN | 68 | 44 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| ABRAHAM ALPC VS MULTIPLE MYELOMA DN | 19 | 14 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED DN | 246 | 180 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP | 390 | 242 | All SZGR 2.0 genes in this pathway |
| MARIADASON REGULATED BY HISTONE ACETYLATION DN | 54 | 30 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS BY HNE AND TBH | 60 | 42 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP | 487 | 303 | All SZGR 2.0 genes in this pathway |
| MCCLUNG COCAIN REWARD 4WK | 75 | 47 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION DN | 287 | 208 | All SZGR 2.0 genes in this pathway |
| KYNG RESPONSE TO H2O2 VIA ERCC6 | 17 | 9 | All SZGR 2.0 genes in this pathway |
| KYNG RESPONSE TO H2O2 | 71 | 42 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| KONDO PROSTATE CANCER HCP WITH H3K27ME3 | 97 | 72 | All SZGR 2.0 genes in this pathway |
| SARRIO EPITHELIAL MESENCHYMAL TRANSITION DN | 154 | 101 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K27ME3 UP | 295 | 149 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL OPTIMAL DEBULKING | 246 | 152 | All SZGR 2.0 genes in this pathway |
| GRADE METASTASIS DN | 45 | 31 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| GRADE COLON VS RECTAL CANCER DN | 56 | 36 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| GU PDEF TARGETS UP | 71 | 49 | All SZGR 2.0 genes in this pathway |
| LINDSTEDT DENDRITIC CELL MATURATION C | 69 | 49 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP | 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 DN | 245 | 150 | All SZGR 2.0 genes in this pathway |
| MUELLER COMMON TARGETS OF AML FUSIONS UP | 14 | 10 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE DN | 181 | 97 | All SZGR 2.0 genes in this pathway |
| ONO AML1 TARGETS DN | 41 | 25 | All SZGR 2.0 genes in this pathway |
| ONO FOXP3 TARGETS DN | 42 | 23 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC UP | 72 | 53 | All SZGR 2.0 genes in this pathway |
| KYNG RESPONSE TO H2O2 VIA ERCC6 DN | 46 | 31 | All SZGR 2.0 genes in this pathway |
| THILLAINADESAN ZNF217 TARGETS UP | 44 | 22 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN | 448 | 282 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |