Gene Page: SUMO1
Summary ?
| GeneID | 7341 |
| Symbol | SUMO1 |
| Synonyms | DAP1|GMP1|OFC10|PIC1|SENP2|SMT3|SMT3C|SMT3H3|UBL1 |
| Description | small ubiquitin-like modifier 1 |
| Reference | MIM:601912|HGNC:HGNC:12502|Ensembl:ENSG00000116030|HPRD:03554|Vega:OTTHUMG00000132839 |
| Gene type | protein-coding |
| Map location | 2q33 |
| Pascal p-value | 0.257 |
| Sherlock p-value | 0.665 |
| Fetal beta | 0.48 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg02592124 | 2 | 203103479 | SUMO1 | 2.47E-10 | -0.011 | 6.18E-7 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs823066 | chr1 | 205771172 | SUMO1 | 7341 | 0.12 | trans | ||
| rs9882137 | chr3 | 29494146 | SUMO1 | 7341 | 0.07 | trans | ||
| rs4491879 | chr3 | 189373908 | SUMO1 | 7341 | 0.11 | trans | ||
| rs4505678 | chr3 | 189374356 | SUMO1 | 7341 | 0.08 | trans | ||
| rs13175086 | chr5 | 174691428 | SUMO1 | 7341 | 0.15 | trans | ||
| rs16894557 | chr6 | 28999825 | SUMO1 | 7341 | 0.1 | trans | ||
| rs1999514 | chr13 | 84147878 | SUMO1 | 7341 | 0.16 | trans | ||
| rs917836 | chr18 | 20053367 | SUMO1 | 7341 | 0.16 | trans | ||
| rs1503026 | chr18 | 70842605 | SUMO1 | 7341 | 0.01 | trans | ||
| rs12326929 | chr18 | 70864592 | SUMO1 | 7341 | 0.01 | trans | ||
| rs12327665 | chr19 | 39622639 | SUMO1 | 7341 | 0.05 | trans |
Section II. Transcriptome annotation
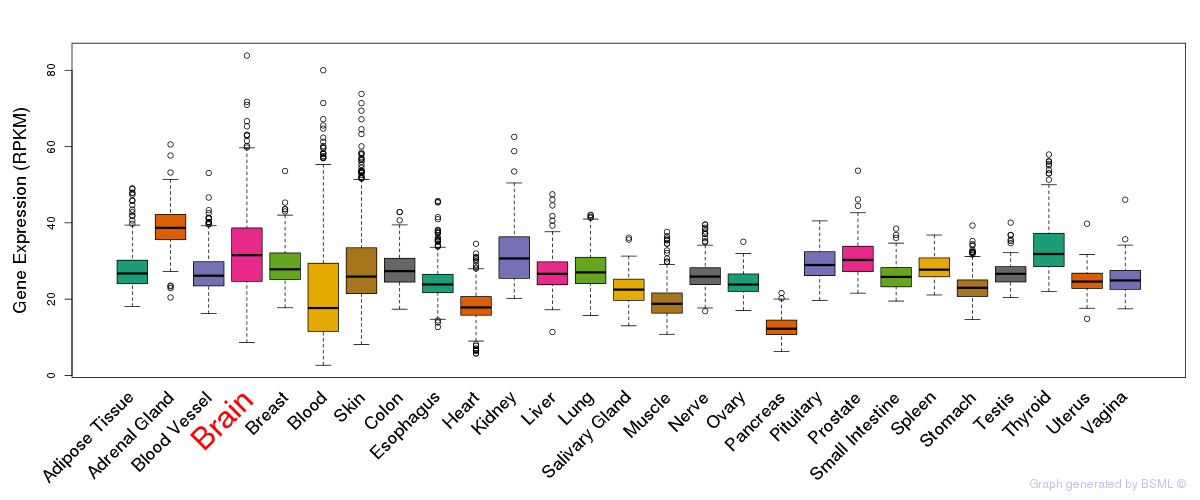
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SLC25A11 | 0.91 | 0.92 |
| C19orf62 | 0.89 | 0.87 |
| PSMD8 | 0.89 | 0.86 |
| MRPL4 | 0.88 | 0.88 |
| WDR45 | 0.88 | 0.87 |
| ANKRD39 | 0.88 | 0.85 |
| MRPL38 | 0.88 | 0.87 |
| APEH | 0.88 | 0.85 |
| TMEM9 | 0.88 | 0.86 |
| STOML2 | 0.87 | 0.89 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.18 | -0.59 | -0.48 |
| AC010300.1 | -0.55 | -0.60 |
| MT-ATP8 | -0.54 | -0.41 |
| AF347015.26 | -0.53 | -0.39 |
| AF347015.8 | -0.53 | -0.37 |
| AF347015.2 | -0.50 | -0.33 |
| AF347015.15 | -0.49 | -0.35 |
| C10orf108 | -0.49 | -0.45 |
| AC100783.1 | -0.48 | -0.41 |
| AF347015.21 | -0.48 | -0.33 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| C11orf65 | MGC33948 | chromosome 11 open reading frame 65 | Two-hybrid | BioGRID | 16189514 |
| CASP8 | ALPS2B | CAP4 | FLICE | FLJ17672 | MACH | MCH5 | MGC78473 | caspase 8, apoptosis-related cysteine peptidase | CASP8 (caspase-8) interacts with SUMO1. | BIND | 15782135 |
| CBX4 | NBP16 | PC2 | hPC2 | chromobox homolog 4 (Pc class homolog, Drosophila) | Pc2 interacts with SUMO1. | BIND | 15592428 |
| CD40 | Bp50 | CDW40 | MGC9013 | TNFRSF5 | p50 | CD40 molecule, TNF receptor superfamily member 5 | Two-hybrid | BioGRID | 8906799 |
| CHD3 | Mi-2a | Mi2-ALPHA | ZFH | chromodomain helicase DNA binding protein 3 | SUMO-1 interacts with CHD3. | BIND | 14609633 |
| CHD3 | Mi-2a | Mi2-ALPHA | ZFH | chromodomain helicase DNA binding protein 3 | - | HPRD | 14609633 |
| CHD3 | Mi-2a | Mi2-ALPHA | ZFH | chromodomain helicase DNA binding protein 3 | Two-hybrid | BioGRID | 10961991 |
| DAXX | BING2 | DAP6 | EAP1 | MGC126245 | MGC126246 | death-domain associated protein | Daxx interacts with sentrin. | BIND | 11112409 |
| DAXX | BING2 | DAP6 | EAP1 | MGC126245 | MGC126246 | death-domain associated protein | - | HPRD,BioGRID | 11112409 |
| DAXX | BING2 | DAP6 | EAP1 | MGC126245 | MGC126246 | death-domain associated protein | Daxx interacts with an unspecified isoform of SUMO1. | BIND | 11948183 |
| DNMT3B | ICF | M.HsaIIIB | DNA (cytosine-5-)-methyltransferase 3 beta | Dnmt3b interacts with SUMO-1. | BIND | 11735126 |
| DNMT3B | ICF | M.HsaIIIB | DNA (cytosine-5-)-methyltransferase 3 beta | - | HPRD,BioGRID | 11735126 |
| FADD | GIG3 | MGC8528 | MORT1 | Fas (TNFRSF6)-associated via death domain | Two-hybrid | BioGRID | 8906799 |
| FAS | ALPS1A | APO-1 | APT1 | CD95 | FAS1 | FASTM | TNFRSF6 | Fas (TNF receptor superfamily, member 6) | Reconstituted Complex Two-hybrid | BioGRID | 8906799 |11112409 |
| FAS | ALPS1A | APO-1 | APT1 | CD95 | FAS1 | FASTM | TNFRSF6 | Fas (TNF receptor superfamily, member 6) | Fas interacts with sentrin. | BIND | 11112409 |
| FAS | ALPS1A | APO-1 | APT1 | CD95 | FAS1 | FASTM | TNFRSF6 | Fas (TNF receptor superfamily, member 6) | - | HPRD | 8906799 |9740801 |
| FASLG | APT1LG1 | CD178 | CD95L | FASL | TNFSF6 | Fas ligand (TNF superfamily, member 6) | - | HPRD,BioGRID | 8906799 |
| HDAC9 | DKFZp779K1053 | HD7 | HDAC | HDAC7 | HDAC7B | HDAC9B | HDAC9FL | HDRP | KIAA0744 | MITR | histone deacetylase 9 | - | HPRD | 12590135 |
| HIPK2 | DKFZp686K02111 | FLJ23711 | PRO0593 | homeodomain interacting protein kinase 2 | SUMO-1 interacts with HIPK2. | BIND | 14609633 |
| HIPK2 | DKFZp686K02111 | FLJ23711 | PRO0593 | homeodomain interacting protein kinase 2 | - | HPRD,BioGRID | 14609633 |
| HIPK3 | DYRK6 | FIST3 | PKY | YAK1 | homeodomain interacting protein kinase 3 | Two-hybrid | BioGRID | 10961991 |
| HTT | HD | IT15 | huntingtin | HD is covalently attached to SUMO-1 via lysine residues. | BIND | 15064418 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | - | HPRD | 10892746 |
| MITF | MI | WS2A | bHLHe32 | microphthalmia-associated transcription factor | - | HPRD,BioGRID | 10694430 |
| NR3C1 | GCCR | GCR | GR | GRL | nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | - | HPRD | 12193561 |
| PIAS1 | DDXBP1 | GBP | GU/RH-II | MGC141878 | MGC141879 | ZMIZ3 | protein inhibitor of activated STAT, 1 | SUMO-1 interacts with PIAS1. | BIND | 14609633 |
| PIAS1 | DDXBP1 | GBP | GU/RH-II | MGC141878 | MGC141879 | ZMIZ3 | protein inhibitor of activated STAT, 1 | - | HPRD,BioGRID | 11583632 |
| PIAS2 | MGC102682 | MIZ1 | PIASX | PIASX-ALPHA | PIASX-BETA | SIZ2 | ZMIZ4 | miz | protein inhibitor of activated STAT, 2 | SUMO-1 interacts with PIASxbeta. | BIND | 14609633 |
| PIAS2 | MGC102682 | MIZ1 | PIASX | PIASX-ALPHA | PIASX-BETA | SIZ2 | ZMIZ4 | miz | protein inhibitor of activated STAT, 2 | Two-hybrid | BioGRID | 14609633 |
| PML | MYL | PP8675 | RNF71 | TRIM19 | promyelocytic leukemia | - | HPRD | 8806687 |11080164 |
| PML | MYL | PP8675 | RNF71 | TRIM19 | promyelocytic leukemia | Affinity Capture-Western in vitro in vivo Two-hybrid | BioGRID | 8806687 |9452416 |11948183 |
| PPM1J | DKFZp434P1514 | FLJ35951 | MGC19531 | MGC90149 | PP2CZ | PP2Czeta | PPP2CZ | protein phosphatase 1J (PP2C domain containing) | - | HPRD | 12633878 |
| RAD51 | BRCC5 | HRAD51 | HsRad51 | HsT16930 | RAD51A | RECA | RAD51 homolog (RecA homolog, E. coli) (S. cerevisiae) | - | HPRD,BioGRID | 8812453 |9891849 |
| RAD52 | - | RAD52 homolog (S. cerevisiae) | Two-hybrid | BioGRID | 8812453 |
| RANBP2 | NUP358 | TRP1 | TRP2 | RAN binding protein 2 | Reconstituted Complex | BioGRID | 12192048 |
| RANBP2 | NUP358 | TRP1 | TRP2 | RAN binding protein 2 | SUMO-1 interacts with RanBP2. | BIND | 15608651 |
| RANGAP1 | Fug1 | KIAA1835 | MGC20266 | SD | Ran GTPase activating protein 1 | - | HPRD | 9019411 |
| RANGAP1 | Fug1 | KIAA1835 | MGC20266 | SD | Ran GTPase activating protein 1 | RanGAP1 interacts with and is covalently modified by SUMO-1. | BIND | 9920803 |
| RANGAP1 | Fug1 | KIAA1835 | MGC20266 | SD | Ran GTPase activating protein 1 | Reconstituted Complex | BioGRID | 12924945 |
| SAE1 | AOS1 | FLJ3091 | HSPC140 | SUA1 | SUMO1 activating enzyme subunit 1 | Reconstituted Complex | BioGRID | 12924945 |
| SAE1 | AOS1 | FLJ3091 | HSPC140 | SUA1 | SUMO1 activating enzyme subunit 1 | - | HPRD | 10187858 |
| SALL1 | HSAL1 | TBS | ZNF794 | sal-like 1 (Drosophila) | - | HPRD,BioGRID | 12200128 |
| SALL1 | HSAL1 | TBS | ZNF794 | sal-like 1 (Drosophila) | SALL1 interacts with SUMO-1. | BIND | 12200128 |
| SENP2 | AXAM2 | DKFZp762A2316 | KIAA1331 | SMT3IP2 | SUMO1/sentrin/SMT3 specific peptidase 2 | - | HPRD | 11997515 |
| SENP2 | AXAM2 | DKFZp762A2316 | KIAA1331 | SMT3IP2 | SUMO1/sentrin/SMT3 specific peptidase 2 | Biochemical Activity | BioGRID | 12192048 |
| SENP6 | FLJ11355 | FLJ11887 | KIAA0389 | KIAA0797 | SSP1 | SUSP1 | SUMO1/sentrin specific peptidase 6 | - | HPRD | 10799485 |
| SLC2A1 | DYT17 | DYT18 | GLUT | GLUT1 | MGC141895 | MGC141896 | PED | solute carrier family 2 (facilitated glucose transporter), member 1 | - | HPRD | 10655495 |
| SMAD4 | DPC4 | JIP | MADH4 | SMAD family member 4 | SMAD4 is sumoylated. | BIND | 15592428 |
| SP100 | DKFZp686E07254 | FLJ00340 | FLJ34579 | SP100 nuclear antigen | Two-hybrid | BioGRID | 16189514 |
| SREBF2 | SREBP2 | bHLHd2 | sterol regulatory element binding transcription factor 2 | - | HPRD | 12615929 |
| STAB2 | DKFZp434E0321 | FEEL-2 | FELE-2 | FELL | FELL-2 | FEX2 | HARE | STAB-2 | stabilin 2 | Biochemical Activity | BioGRID | 14701874 |
| SUMO1 | DAP-1 | GMP1 | OFC10 | PIC1 | SENP2 | SMT3 | SMT3C | SMT3H3 | SUMO-1 | UBL1 | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | Two-hybrid | BioGRID | 10961991 |11112409 |
| SUMO1 | DAP-1 | GMP1 | OFC10 | PIC1 | SENP2 | SMT3 | SMT3C | SMT3H3 | SUMO-1 | UBL1 | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | Sentrin interacts with sentrin to form a homodimer. | BIND | 11112409 |
| TDG | - | thymine-DNA glycosylase | - | HPRD,BioGRID | 11889051 |
| TNFRSF1A | CD120a | FPF | MGC19588 | TBP1 | TNF-R | TNF-R-I | TNF-R55 | TNFAR | TNFR1 | TNFR55 | TNFR60 | p55 | p55-R | p60 | tumor necrosis factor receptor superfamily, member 1A | Reconstituted Complex Two-hybrid | BioGRID | 8906799 |10187798 |
| TOE1 | FLJ13949 | target of EGR1, member 1 (nuclear) | Two-hybrid | BioGRID | 16169070 |
| TOP1 | TOPI | topoisomerase (DNA) I | - | HPRD,BioGRID | 11709553 |
| TOP2A | TOP2 | TP2A | topoisomerase (DNA) II alpha 170kDa | - | HPRD,BioGRID | 10862613 |12832072 |
| TOP2B | TOPIIB | top2beta | topoisomerase (DNA) II beta 180kDa | - | HPRD,BioGRID | 10862613 |
| TOPORS | LUN | P53BP3 | RP31 | TP53BPL | topoisomerase I binding, arginine/serine-rich | - | HPRD | 11842245 |
| TOPORS | LUN | P53BP3 | RP31 | TP53BPL | topoisomerase I binding, arginine/serine-rich | Two-hybrid | BioGRID | 14516784 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | Two-hybrid | BioGRID | 10961991 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | p53 interacts with SUMO-1. This interaction was modelled on a demonstrated interaction between p53 from human and SUMO-1 from an unspecified species. | BIND | 10788439 |
| TP73 | P73 | tumor protein p73 | Two-hybrid | BioGRID | 10961991 |
| TTRAP | AD022 | EAP2 | MGC111021 | MGC9099 | dJ30M3.3 | TRAF and TNF receptor associated protein | SUMO-1 interacts with TTRAP. | BIND | 14609633 |
| TTRAP | AD022 | EAP2 | MGC111021 | MGC9099 | dJ30M3.3 | TRAF and TNF receptor associated protein | Two-hybrid | BioGRID | 14609633 |
| UBA2 | ARX | FLJ13058 | HRIHFB2115 | SAE2 | ubiquitin-like modifier activating enzyme 2 | SAE2 and an unspecified isoform of SUMO-1 form thioester bond. This interaction was modeled on a demonstrated interaction between SAE2 from an unspecified species and SUMO-1 from an unspecified species. | BIND | 15546615 |
| UBA2 | ARX | FLJ13058 | HRIHFB2115 | SAE2 | ubiquitin-like modifier activating enzyme 2 | - | HPRD,BioGRID | 10217437 |
| UBE2I | C358B7.1 | P18 | UBC9 | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | - | HPRD,BioGRID | 12924945 |
| UBE2I | C358B7.1 | P18 | UBC9 | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | Ubc9 interacts with SUMO1. | BIND | 15592428 |
| UBE2I | C358B7.1 | P18 | UBC9 | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | hUbc9 interacts with SUMO-1. | BIND | 9920803 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA PML PATHWAY | 17 | 12 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PARKIN PATHWAY | 13 | 8 | All SZGR 2.0 genes in this pathway |
| PID HDAC CLASSII PATHWAY | 34 | 27 | All SZGR 2.0 genes in this pathway |
| PID HDAC CLASSI PATHWAY | 66 | 50 | All SZGR 2.0 genes in this pathway |
| PID RANBP2 PATHWAY | 11 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF IFNG SIGNALING | 14 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERFERON GAMMA SIGNALING | 63 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERFERON SIGNALING | 159 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| WILCOX RESPONSE TO PROGESTERONE UP | 152 | 90 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| CHOI ATL CHRONIC VS ACUTE DN | 18 | 10 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS UP | 372 | 227 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| OUELLET OVARIAN CANCER INVASIVE VS LMP UP | 117 | 85 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN MELANOMA RELAPSE UP | 61 | 25 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS UP | 344 | 180 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION DN | 517 | 309 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS UP | 424 | 268 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO DN | 200 | 112 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST DN | 309 | 206 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA SUBGROUPS | 30 | 20 | All SZGR 2.0 genes in this pathway |
| KAAB FAILED HEART ATRIUM DN | 141 | 99 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA CANCER | 83 | 52 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC | 420 | 269 | All SZGR 2.0 genes in this pathway |
| SHAFFER IRF4 TARGETS IN MYELOMA VS MATURE B LYMPHOCYTE | 101 | 76 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS UP | 518 | 299 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE UP | 442 | 263 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S2 | 115 | 74 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING DN | 225 | 124 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN | 253 | 192 | All SZGR 2.0 genes in this pathway |
| WONG EMBRYONIC STEM CELL CORE | 335 | 193 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 10 | 69 | 38 | All SZGR 2.0 genes in this pathway |
| NAKAMURA ADIPOGENESIS LATE UP | 104 | 67 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |