Gene Page: YY1
Summary ?
| GeneID | 7528 |
| Symbol | YY1 |
| Synonyms | DELTA|INO80S|NF-E1|UCRBP|YIN-YANG-1 |
| Description | YY1 transcription factor |
| Reference | MIM:600013|HGNC:HGNC:12856|HPRD:02482| |
| Gene type | protein-coding |
| Map location | 14q |
| Pascal p-value | 0.501 |
| Sherlock p-value | 3.053E-4 |
| Fetal beta | 0.117 |
| eGene | Nucleus accumbens basal ganglia Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0278 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17145698 | chrX | 40218345 | YY1 | 7528 | 0.18 | trans |
Section II. Transcriptome annotation
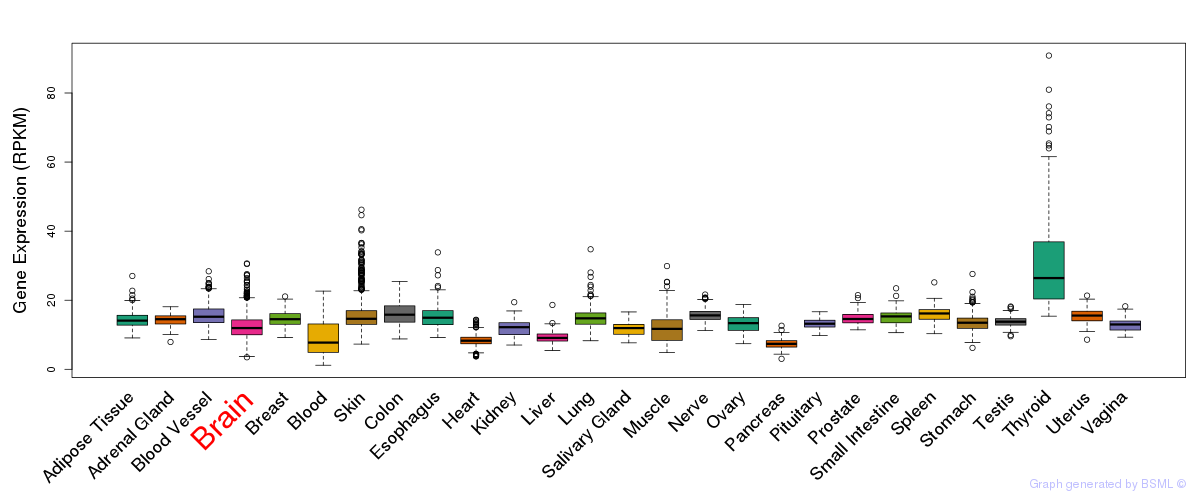
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MARK3 | 0.94 | 0.95 |
| ZNF280D | 0.93 | 0.94 |
| LEO1 | 0.93 | 0.95 |
| ZNF711 | 0.92 | 0.95 |
| REV1 | 0.92 | 0.94 |
| SCAPER | 0.91 | 0.94 |
| TTC17 | 0.91 | 0.93 |
| SR140 | 0.91 | 0.94 |
| C10orf137 | 0.91 | 0.94 |
| ZFP28 | 0.91 | 0.95 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.82 | -0.88 |
| MT-CO2 | -0.81 | -0.88 |
| AF347015.27 | -0.81 | -0.87 |
| AF347015.33 | -0.79 | -0.86 |
| FXYD1 | -0.79 | -0.86 |
| IFI27 | -0.79 | -0.87 |
| MT-CYB | -0.78 | -0.85 |
| AF347015.8 | -0.78 | -0.87 |
| HIGD1B | -0.77 | -0.86 |
| HLA-F | -0.77 | -0.79 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | TAS | 1946405 | |
| GO:0003713 | transcription coactivator activity | TAS | 1946405 | |
| GO:0003714 | transcription corepressor activity | TAS | 1655281 | |
| GO:0005515 | protein binding | IPI | 9016636 | |
| GO:0008270 | zinc ion binding | TAS | 1655281 | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006357 | regulation of transcription from RNA polymerase II promoter | TAS | 1655281 | |
| GO:0006350 | transcription | IEA | - | |
| GO:0009952 | anterior/posterior pattern formation | IEA | - | |
| GO:0048593 | camera-type eye morphogenesis | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005667 | transcription factor complex | IEA | - | |
| GO:0005737 | cytoplasm | IDA | 18029348 | |
| GO:0005886 | plasma membrane | IDA | 18029348 | |
| GO:0016363 | nuclear matrix | IEA | - | |
| GO:0031519 | PcG protein complex | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ATF2 | CRE-BP1 | CREB2 | HB16 | MGC111558 | TREB7 | activating transcription factor 2 | YY1 interacts with ATF-2. | BIND | 7769693 |
| ATF6 | ATF6A | activating transcription factor 6 | - | HPRD,BioGRID | 10866666 |
| ATF7 | ATFA | MGC57182 | activating transcription factor 7 | YY1 interacts with ATFa2. | BIND | 7769693 |
| ATF7 | ATFA | MGC57182 | activating transcription factor 7 | YY1 interacts with ATFa3. | BIND | 7769693 |
| CDKN2A | ARF | CDK4I | CDKN2 | CMM2 | INK4 | INK4a | MLM | MTS1 | TP16 | p14 | p14ARF | p16 | p16INK4 | p16INK4a | p19 | cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | YY1 interacts with p14ARF. | BIND | 15210108 |
| CREB1 | CREB | MGC9284 | cAMP responsive element binding protein 1 | YY1 interacts with CREB. | BIND | 7769693 |
| DNAJB4 | DNAJW | DjB4 | HLJ1 | DnaJ (Hsp40) homolog, subfamily B, member 4 | YY1 interacts with HLJ1 YY1-binding site. | BIND | 15782117 |
| E2F2 | E2F-2 | E2F transcription factor 2 | - | HPRD | 12411495 |
| E2F3 | DKFZp686C18211 | E2F-3 | KIAA0075 | MGC104598 | E2F transcription factor 3 | - | HPRD | 12411495 |
| EED | HEED | WAIT1 | embryonic ectoderm development | YY1 interacts with EED. | BIND | 14610174 |
| EED | HEED | WAIT1 | embryonic ectoderm development | EED interacts with YY1. | BIND | 11158321 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | Affinity Capture-Western Biochemical Activity Reconstituted Complex Two-hybrid | BioGRID | 7758944 |11486036 |
| EZH2 | ENX-1 | EZH1 | KMT6 | MGC9169 | enhancer of zeste homolog 2 (Drosophila) | EED interacts with EZH2. | BIND | 14610174 |
| FKBP1A | FKBP-12 | FKBP1 | FKBP12 | FKBP12C | PKC12 | PKCI2 | PPIASE | FK506 binding protein 1A, 12kDa | Two-hybrid | BioGRID | 7541038 |
| FKBP3 | FKBP-25 | PPIase | FK506 binding protein 3, 25kDa | Affinity Capture-Western Reconstituted Complex | BioGRID | 11532945 |
| GTF2I | BAP-135 | BAP135 | BTKAP1 | DIWS | FLJ38776 | FLJ56355 | IB291 | SPIN | TFII-I | WBS | WBSCR6 | general transcription factor II, i | - | HPRD | 11287625 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | Reconstituted Complex | BioGRID | 9346952 |
| HDAC2 | RPD3 | YAF1 | histone deacetylase 2 | YY1 interacts with RPD3. This interaction was modelled on a demonstrated interaction between human YY1 and mouse RPD3. | BIND | 8917507 |
| HDAC2 | RPD3 | YAF1 | histone deacetylase 2 | - | HPRD,BioGRID | 11486036 |
| HDAC3 | HD3 | RPD3 | RPD3-2 | histone deacetylase 3 | - | HPRD,BioGRID | 9346952 |
| KAT2B | CAF | P | P/CAF | PCAF | K(lysine) acetyltransferase 2B | Biochemical Activity | BioGRID | 11486036 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | YY1 interacts with Hdm2. | BIND | 15210108 |
| MTA2 | DKFZp686F2281 | MTA1L1 | PID | metastasis associated 1 family, member 2 | Reconstituted Complex | BioGRID | 12920132 |
| MYC | bHLHe39 | c-Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | - | HPRD,BioGRID | 8266081 |
| NOTCH1 | TAN1 | hN1 | Notch homolog 1, translocation-associated (Drosophila) | - | HPRD,BioGRID | 12913000 |
| RYBP | AAP1 | DEDAF | YEAF1 | RING1 and YY1 binding protein | - | HPRD,BioGRID | 10369680 |
| SAP30 | - | Sin3A-associated protein, 30kDa | - | HPRD,BioGRID | 12788099 |
| SP1 | - | Sp1 transcription factor | Affinity Capture-Western | BioGRID | 10224053 |
| SP1 | - | Sp1 transcription factor | YY1 interacts with Sp1. | BIND | 8327494 |
| SREBF1 | SREBP-1c | SREBP1 | bHLHd1 | sterol regulatory element binding transcription factor 1 | Reconstituted Complex | BioGRID | 10224053 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | YY1 interacts with p53. | BIND | 15210108 |
| YAF2 | MGC41856 | YY1 associated factor 2 | - | HPRD,BioGRID | 9016636 |
| ZRANB2 | DKFZp686J1831 | DKFZp686N09117 | FLJ41119 | ZIS | ZIS1 | ZIS2 | ZNF265 | zinc finger, RAN-binding domain containing 2 | - | HPRD | 11448987 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID NOTCH PATHWAY | 59 | 49 | All SZGR 2.0 genes in this pathway |
| PID E2F PATHWAY | 74 | 48 | All SZGR 2.0 genes in this pathway |
| PID HDAC CLASSI PATHWAY | 66 | 50 | All SZGR 2.0 genes in this pathway |
| PID MTOR 4PATHWAY | 69 | 55 | All SZGR 2.0 genes in this pathway |
| PID P53 REGULATION PATHWAY | 59 | 50 | All SZGR 2.0 genes in this pathway |
| PID HES HEY PATHWAY | 48 | 39 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| DAVICIONI TARGETS OF PAX FOXO1 FUSIONS DN | 68 | 49 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS TOP50 UP | 38 | 27 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 UP | 418 | 263 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER MYC TARGETS REPRESSED BY SERUM | 159 | 93 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| LOCKWOOD AMPLIFIED IN LUNG CANCER | 214 | 139 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 240 HELA | 60 | 43 | All SZGR 2.0 genes in this pathway |
| CHEOK RESPONSE TO MERCAPTOPURINE AND LD MTX UP | 10 | 6 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND DN | 225 | 163 | All SZGR 2.0 genes in this pathway |
| FAELT B CLL WITH VH REARRANGEMENTS DN | 48 | 26 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION NEOCORTEX UP | 83 | 66 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C2 | 54 | 39 | All SZGR 2.0 genes in this pathway |
| MARIADASON REGULATED BY HISTONE ACETYLATION DN | 54 | 30 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| HENDRICKS SMARCA4 TARGETS DN | 53 | 34 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 10HR UP | 101 | 69 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D8 | 40 | 29 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| CROONQUIST STROMAL STIMULATION DN | 13 | 7 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| WONG EMBRYONIC STEM CELL CORE | 335 | 193 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 1HR DN | 222 | 147 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE M G1 | 148 | 95 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BAKKER FOXO3 TARGETS DN | 187 | 109 | All SZGR 2.0 genes in this pathway |
| WANG TNF TARGETS | 24 | 17 | All SZGR 2.0 genes in this pathway |
| YUAN ZNF143 PARTNERS | 22 | 15 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-141/200a | 669 | 675 | 1A | hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG |
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| miR-142-5p | 296 | 302 | 1A | hsa-miR-142-5p | CAUAAAGUAGAAAGCACUAC |
| miR-181 | 789 | 795 | 1A | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-186 | 293 | 300 | 1A,m8 | hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU |
| hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU | ||||
| hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU | ||||
| miR-29 | 774 | 780 | m8 | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
| miR-34/449 | 720 | 726 | 1A | hsa-miR-34abrain | UGGCAGUGUCUUAGCUGGUUGUU |
| hsa-miR-34c | AGGCAGUGUAGUUAGCUGAUUGC | ||||
| hsa-miR-449 | UGGCAGUGUAUUGUUAGCUGGU | ||||
| hsa-miR-449b | AGGCAGUGUAUUGUUAGCUGGC | ||||
| miR-381 | 605 | 611 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU | ||||
| miR-384 | 90 | 96 | m8 | hsa-miR-384 | AUUCCUAGAAAUUGUUCAUA |
| miR-410 | 546 | 553 | 1A,m8 | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| miR-433-3p | 269 | 275 | 1A | hsa-miR-433brain | AUCAUGAUGGGCUCCUCGGUGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.