Gene Page: CAMK2G
Summary ?
| GeneID | 818 |
| Symbol | CAMK2G |
| Synonyms | CAMK|CAMK-II|CAMKG |
| Description | calcium/calmodulin-dependent protein kinase II gamma |
| Reference | MIM:602123|HGNC:HGNC:1463|Ensembl:ENSG00000148660|HPRD:03672|Vega:OTTHUMG00000018492 |
| Gene type | protein-coding |
| Map location | 10q22 |
| Pascal p-value | 0.601 |
| Sherlock p-value | 0.214 |
| Fetal beta | -0.991 |
| DMG | 2 (# studies) |
| eGene | Nucleus accumbens basal ganglia Myers' cis & trans |
| Support | INTRACELLULAR SIGNAL TRANSDUCTION SEROTONIN G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_clathrin G2Cdb.human_Synaptosome G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 3 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 3 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.1968 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg10168709 | 10 | 75599394 | CAMK2G | 5.02E-6 | 0.547 | 0.01 | DMG:Wockner_2014 |
| cg06022113 | 10 | 75616203 | CAMK2G | 9.09E-5 | 0.405 | 0.027 | DMG:Wockner_2014 |
| cg19287139 | 10 | 75634631 | CAMK2G | 2.09E-8 | -0.019 | 7.15E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs9571716 | chr13 | 67711344 | CAMK2G | 818 | 0.19 | trans |
Section II. Transcriptome annotation
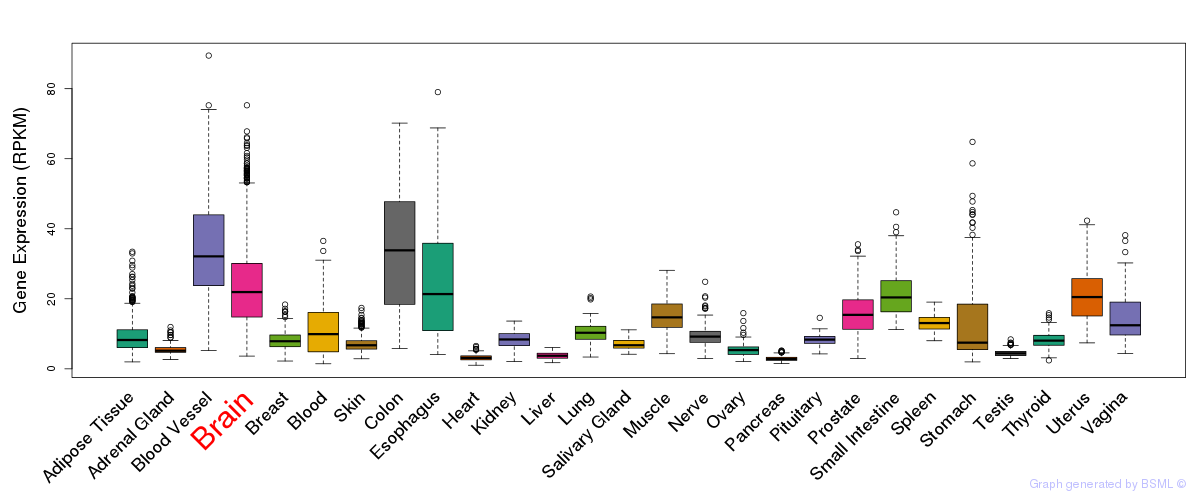
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GNG5 | 0.74 | 0.67 |
| HMG20B | 0.71 | 0.63 |
| TP53I13 | 0.70 | 0.71 |
| PECI | 0.69 | 0.61 |
| TGIF1 | 0.69 | 0.42 |
| RFXANK | 0.69 | 0.54 |
| PAX6 | 0.69 | 0.65 |
| CLIC1 | 0.67 | 0.45 |
| CDKN2C | 0.65 | 0.72 |
| AKR7A2 | 0.64 | 0.62 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CACNA1A | -0.39 | -0.47 |
| ARHGAP20 | -0.38 | -0.48 |
| FBXW7 | -0.38 | -0.46 |
| DOCK9 | -0.37 | -0.47 |
| HIVEP2 | -0.36 | -0.49 |
| LRFN2 | -0.36 | -0.47 |
| RALYL | -0.36 | -0.54 |
| RTN4RL1 | -0.36 | -0.46 |
| FRMPD4 | -0.35 | -0.43 |
| MEF2C | -0.35 | -0.48 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005516 | calmodulin binding | NAS | 9060999 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0005524 | ATP binding | NAS | - | |
| GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity | NAS | 9060999 | |
| GO:0004674 | protein serine/threonine kinase activity | IEA | - | |
| GO:0004683 | calmodulin-dependent protein kinase activity | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0016301 | kinase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000082 | G1/S transition of mitotic cell cycle | IEA | - | |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0006468 | protein amino acid phosphorylation | NAS | - | |
| GO:0007165 | signal transduction | NAS | 8449910 | |
| GO:0006816 | calcium ion transport | IEA | - | |
| GO:0030073 | insulin secretion | NAS | 9240463 | |
| GO:0046777 | protein amino acid autophosphorylation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005575 | cellular_component | ND | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ERBB SIGNALING PATHWAY | 87 | 71 | All SZGR 2.0 genes in this pathway |
| KEGG CALCIUM SIGNALING PATHWAY | 178 | 134 | All SZGR 2.0 genes in this pathway |
| KEGG OOCYTE MEIOSIS | 114 | 79 | All SZGR 2.0 genes in this pathway |
| KEGG WNT SIGNALING PATHWAY | 151 | 112 | All SZGR 2.0 genes in this pathway |
| KEGG LONG TERM POTENTIATION | 70 | 57 | All SZGR 2.0 genes in this pathway |
| KEGG NEUROTROPHIN SIGNALING PATHWAY | 126 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG OLFACTORY TRANSDUCTION | 389 | 85 | All SZGR 2.0 genes in this pathway |
| KEGG GNRH SIGNALING PATHWAY | 101 | 77 | All SZGR 2.0 genes in this pathway |
| KEGG MELANOGENESIS | 102 | 80 | All SZGR 2.0 genes in this pathway |
| KEGG GLIOMA | 65 | 56 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BIOPEPTIDES PATHWAY | 45 | 35 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CACAM PATHWAY | 16 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PGC1A PATHWAY | 26 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA STATHMIN PATHWAY | 19 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CREB PATHWAY | 27 | 25 | All SZGR 2.0 genes in this pathway |
| ST WNT CA2 CYCLIC GMP PATHWAY | 20 | 14 | All SZGR 2.0 genes in this pathway |
| PID BCR 5PATHWAY | 65 | 50 | All SZGR 2.0 genes in this pathway |
| PID IFNG PATHWAY | 40 | 34 | All SZGR 2.0 genes in this pathway |
| PID NCADHERIN PATHWAY | 33 | 32 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER 20Q13 AMPLIFICATION DN | 180 | 101 | All SZGR 2.0 genes in this pathway |
| NAGASHIMA NRG1 SIGNALING UP | 176 | 123 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 DN | 378 | 231 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN UP | 239 | 157 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| JOHANSSON BRAIN CANCER EARLY VS LATE DN | 45 | 35 | All SZGR 2.0 genes in this pathway |
| SHIPP DLBCL CURED VS FATAL DN | 45 | 30 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND DN | 225 | 163 | All SZGR 2.0 genes in this pathway |
| JIANG AGING HYPOTHALAMUS DN | 40 | 31 | All SZGR 2.0 genes in this pathway |
| LEE AGING CEREBELLUM DN | 86 | 66 | All SZGR 2.0 genes in this pathway |
| ULE SPLICING VIA NOVA2 | 43 | 38 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 24HR UP | 148 | 96 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP | 578 | 341 | All SZGR 2.0 genes in this pathway |
| AMBROSINI FLAVOPIRIDOL TREATMENT TP53 | 109 | 63 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 30MIN UP | 56 | 38 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA NEURAL | 129 | 85 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS NOT VIA AKT1 DN | 88 | 61 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-10 | 1698 | 1704 | m8 | hsa-miR-10a | UACCCUGUAGAUCCGAAUUUGUG |
| hsa-miR-10b | UACCCUGUAGAACCGAAUUUGU | ||||
| miR-103/107 | 1115 | 1121 | 1A | hsa-miR-103brain | AGCAGCAUUGUACAGGGCUAUGA |
| hsa-miR-107brain | AGCAGCAUUGUACAGGGCUAUCA | ||||
| hsa-miR-103brain | AGCAGCAUUGUACAGGGCUAUGA | ||||
| hsa-miR-107brain | AGCAGCAUUGUACAGGGCUAUCA | ||||
| miR-153 | 1067 | 1073 | m8 | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-181 | 1935 | 1941 | m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-194 | 1962 | 1968 | 1A | hsa-miR-194 | UGUAACAGCAACUCCAUGUGGA |
| miR-219 | 1232 | 1238 | m8 | hsa-miR-219brain | UGAUUGUCCAAACGCAAUUCU |
| miR-29 | 684 | 690 | m8 | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
| hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU | ||||
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
| miR-324-5p | 767 | 773 | 1A | hsa-miR-324-5p | CGCAUCCCCUAGGGCAUUGGUGU |
| miR-33 | 774 | 780 | m8 | hsa-miR-33 | GUGCAUUGUAGUUGCAUUG |
| hsa-miR-33b | GUGCAUUGCUGUUGCAUUGCA | ||||
| hsa-miR-33 | GUGCAUUGUAGUUGCAUUG | ||||
| hsa-miR-33b | GUGCAUUGCUGUUGCAUUGCA | ||||
| miR-338 | 1227 | 1233 | 1A | hsa-miR-338brain | UCCAGCAUCAGUGAUUUUGUUGA |
| miR-495 | 1960 | 1966 | 1A | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
| hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU | ||||
| miR-543 | 1936 | 1942 | m8 | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
| miR-544 | 690 | 696 | m8 | hsa-miR-544 | AUUCUGCAUUUUUAGCAAGU |
| hsa-miR-544 | AUUCUGCAUUUUUAGCAAGU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.