Gene Page: CDC20
Summary ?
| GeneID | 991 |
| Symbol | CDC20 |
| Synonyms | CDC20A|bA276H19.3|p55CDC |
| Description | cell division cycle 20 |
| Reference | MIM:603618|HGNC:HGNC:1723|Ensembl:ENSG00000117399|HPRD:04686|Vega:OTTHUMG00000007420 |
| Gene type | protein-coding |
| Map location | 1p34.1 |
| Pascal p-value | 4.865E-5 |
| Sherlock p-value | 0.91 |
| Fetal beta | 2.407 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0167 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2059443 | chr2 | 34308478 | CDC20 | 991 | 0.19 | trans | ||
| rs5003961 | chr7 | 78072443 | CDC20 | 991 | 0.17 | trans | ||
| rs4515471 | chr7 | 78072585 | CDC20 | 991 | 0.17 | trans | ||
| rs10259327 | chr7 | 78074924 | CDC20 | 991 | 0.17 | trans | ||
| rs6991445 | chr8 | 75032923 | CDC20 | 991 | 0.08 | trans | ||
| rs16924550 | chr12 | 22174951 | CDC20 | 991 | 0.1 | trans | ||
| rs11848694 | chr14 | 34320757 | CDC20 | 991 | 0.01 | trans | ||
| rs16979902 | chrX | 140953782 | CDC20 | 991 | 0.1 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
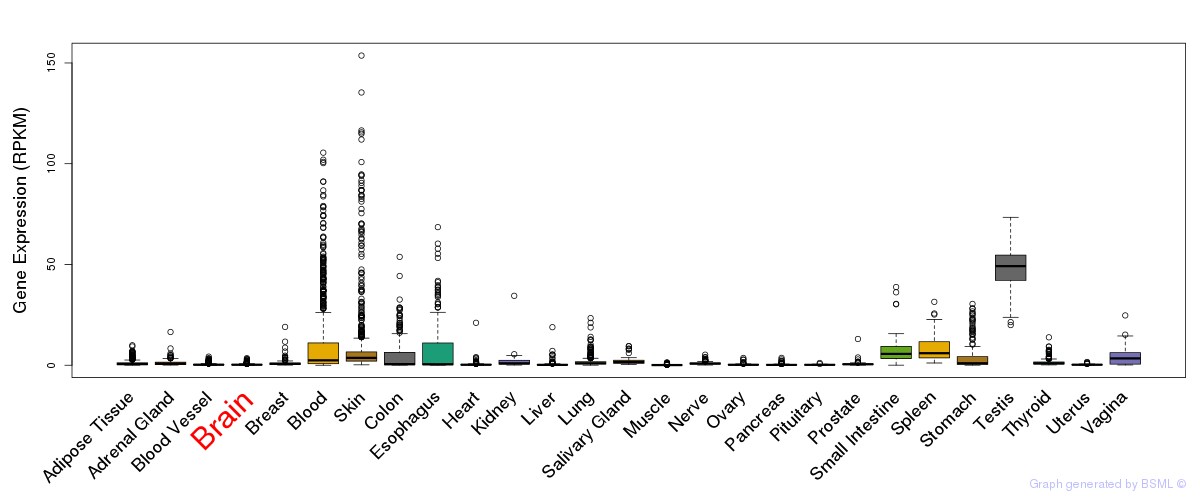
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SLC25A46 | 0.90 | 0.90 |
| PJA2 | 0.89 | 0.91 |
| SLC30A9 | 0.89 | 0.90 |
| C5orf22 | 0.89 | 0.90 |
| PPP6C | 0.89 | 0.90 |
| ACTR2 | 0.89 | 0.90 |
| IPO11 | 0.89 | 0.91 |
| OPA1 | 0.89 | 0.90 |
| KIAA1012 | 0.88 | 0.90 |
| CUL5 | 0.88 | 0.91 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HIGD1B | -0.65 | -0.66 |
| FXYD1 | -0.64 | -0.66 |
| GSDMD | -0.62 | -0.65 |
| MT-CO2 | -0.62 | -0.62 |
| AF347015.21 | -0.60 | -0.59 |
| IFI27 | -0.60 | -0.64 |
| AF347015.2 | -0.60 | -0.58 |
| AF347015.33 | -0.59 | -0.60 |
| AF347015.8 | -0.59 | -0.60 |
| AC098691.2 | -0.59 | -0.61 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 11030144 |14743218 |17353931 |17719540 |18022367 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006511 | ubiquitin-dependent protein catabolic process | TAS | 9682218 | |
| GO:0007049 | cell cycle | TAS | 9682218 | |
| GO:0007067 | mitosis | IEA | - | |
| GO:0031145 | anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | EXP | 11285280 | |
| GO:0051301 | cell division | IEA | - | |
| GO:0051436 | negative regulation of ubiquitin-protein ligase activity during mitotic cell cycle | EXP | 14593737 | |
| GO:0051437 | positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | EXP | 12791267 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005819 | spindle | TAS | 7513050 | |
| GO:0005829 | cytosol | EXP | 11285280 |11535616 |11742988 |12070128 |15469984 | |
| GO:0005654 | nucleoplasm | EXP | 12791267 |15469984 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ANAPC5 | APC5 | anaphase promoting complex subunit 5 | Reconstituted Complex | BioGRID | 12956947 |
| ANAPC7 | APC7 | anaphase promoting complex subunit 7 | Reconstituted Complex | BioGRID | 12956947 |
| APC | BTPS2 | DP2 | DP2.5 | DP3 | GS | adenomatous polyposis coli | - | HPRD | 9637688 |9811605 |
| AURKA | AIK | ARK1 | AURA | AURORA2 | BTAK | MGC34538 | STK15 | STK6 | STK7 | aurora kinase A | - | HPRD,BioGRID | 10377410 |
| BUB1 | BUB1A | BUB1L | hBUB1 | budding uninhibited by benzimidazoles 1 homolog (yeast) | Bub1 interacts with Cdc20. | BIND | 15525512 |
| BUB1B | BUB1beta | BUBR1 | Bub1A | MAD3L | SSK1 | hBUBR1 | budding uninhibited by benzimidazoles 1 homolog beta (yeast) | BubR1 interacts with Cdc20. | BIND | 15525512 |
| BUB1B | BUB1beta | BUBR1 | Bub1A | MAD3L | SSK1 | hBUBR1 | budding uninhibited by benzimidazoles 1 homolog beta (yeast) | - | HPRD,BioGRID | 11030144 |11274370 |11535616 |11907259 |
| CCDC59 | BR22 | DKFZp686K1021 | FLJ10294 | HSPC128 | TAP26 | coiled-coil domain containing 59 | Affinity Capture-MS | BioGRID | 17353931 |
| CCNA2 | CCN1 | CCNA | cyclin A2 | - | HPRD,BioGRID | 10679238 |
| CCNB1 | CCNB | cyclin B1 | Cyclin B interacts with Cdc20. | BIND | 15916961 |
| CDC16 | APC6 | cell division cycle 16 homolog (S. cerevisiae) | - | HPRD,BioGRID | 9628895 |12196507 |
| CDC27 | APC3 | CDC27Hs | D0S1430E | D17S978E | HNUC | cell division cycle 27 homolog (S. cerevisiae) | Affinity Capture-Western Reconstituted Complex | BioGRID | 9628895 |9736712 |9811605 |12196507 |12956947 |14561775 |
| CDC27 | APC3 | CDC27Hs | D0S1430E | D17S978E | HNUC | cell division cycle 27 homolog (S. cerevisiae) | - | HPRD | 9736712 |12196507 |
| FBXO5 | EMI1 | FBX5 | Fbxo31 | F-box protein 5 | - | HPRD,BioGRID | 11389834 |11976684 |11988738 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | Affinity Capture-Western Co-purification | BioGRID | 15388328 |
| HDAC2 | RPD3 | YAF1 | histone deacetylase 2 | Affinity Capture-Western Co-purification | BioGRID | 15388328 |
| KRT31 | HA1 | Ha-1 | KRTHA1 | MGC138630 | hHa1 | keratin 31 | Affinity Capture-MS | BioGRID | 17353931 |
| KRT32 | HA2 | HKA2 | KRTHA2 | hHa2 | keratin 32 | Affinity Capture-MS | BioGRID | 17353931 |
| KRT33A | HA3I | Ha-3I | KRTHA3A | Krt1-3 | hHa3-I | keratin 33A | Affinity Capture-MS | BioGRID | 17353931 |
| KRT85 | HB5 | Hb-5 | KRTHB5 | hHb5 | keratin 85 | Affinity Capture-MS | BioGRID | 17353931 |
| KRTAP19-5 | KAP19.5 | keratin associated protein 19-5 | Affinity Capture-MS | BioGRID | 17353931 |
| KRTAP9-2 | KAP9.2 | KRTAP9.2 | MGC130013 | keratin associated protein 9-2 | Affinity Capture-MS | BioGRID | 17353931 |
| KRTAP9-3 | KAP9.3 | KRTAP9.3 | keratin associated protein 9-3 | Affinity Capture-MS | BioGRID | 17353931 |
| MAD2L1 | HSMAD2 | MAD2 | MAD2 mitotic arrest deficient-like 1 (yeast) | - | HPRD | 9628895 |9637688 |9736712 |10700282 |11804586 |
| MAD2L1 | HSMAD2 | MAD2 | MAD2 mitotic arrest deficient-like 1 (yeast) | MAD2 interacts with p55CDC. | BIND | 12456649 |
| MAD2L1 | HSMAD2 | MAD2 | MAD2 mitotic arrest deficient-like 1 (yeast) | MAD2L1 (Mad2) interacts with Cdc20. | BIND | 15694304 |
| MAD2L1 | HSMAD2 | MAD2 | MAD2 mitotic arrest deficient-like 1 (yeast) | Mad2 interacts with Cdc20. | BIND | 15525512 |
| MAD2L1 | HSMAD2 | MAD2 | MAD2 mitotic arrest deficient-like 1 (yeast) | Affinity Capture-Western Co-crystal Structure Reconstituted Complex Two-hybrid | BioGRID | 9628895 |9637688 |9736712 |10700282 |11438673 |11707408 |12196507 |14561775 |14607335 |
| MAD2L2 | MAD2B | REV7 | MAD2 mitotic arrest deficient-like 2 (yeast) | Reconstituted Complex | BioGRID | 11459826 |
| PTTG1 | EAP1 | HPTTG | MGC126883 | MGC138276 | PTTG | TUTR1 | pituitary tumor-transforming 1 | - | HPRD | 11553328 |
| PTTG1 | EAP1 | HPTTG | MGC126883 | MGC138276 | PTTG | TUTR1 | pituitary tumor-transforming 1 | Securin interacts with Cdc20. This interaction was modelled on a demonstrated interaction between human Securin and Xenopus Cdc20. | BIND | 15916961 |
| SOSTDC1 | CDA019 | DKFZp564D206 | ECTODIN | USAG1 | sclerostin domain containing 1 | - | HPRD | 15280373 |
| SPATC1 | MGC61633 | SPATA15 | SPERIOLIN | spermatogenesis and centriole associated 1 | - | HPRD,BioGRID | 15280373 |
| YARS2 | CGI-04 | FLJ13995 | TYRRS | mt-TyrRS | tyrosyl-tRNA synthetase 2, mitochondrial | Affinity Capture-MS | BioGRID | 17353931 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CELL CYCLE | 128 | 84 | All SZGR 2.0 genes in this pathway |
| KEGG OOCYTE MEIOSIS | 114 | 79 | All SZGR 2.0 genes in this pathway |
| KEGG UBIQUITIN MEDIATED PROTEOLYSIS | 138 | 98 | All SZGR 2.0 genes in this pathway |
| PID PLK1 PATHWAY | 46 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | 24 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE CHECKPOINTS | 124 | 70 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF MITOTIC CELL CYCLE | 85 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC M M G1 PHASES | 172 | 98 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPLICATION | 192 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | 72 | 38 | All SZGR 2.0 genes in this pathway |
| REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | 73 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC PROMETAPHASE | 87 | 51 | All SZGR 2.0 genes in this pathway |
| REACTOME APC C CDC20 MEDIATED DEGRADATION OF CYCLIN B | 26 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | 51 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | 22 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS I MHC MEDIATED ANTIGEN PROCESSING PRESENTATION | 251 | 156 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | 212 | 129 | All SZGR 2.0 genes in this pathway |
| REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | 28 | 12 | All SZGR 2.0 genes in this pathway |
| NAKAMURA CANCER MICROENVIRONMENT DN | 46 | 29 | All SZGR 2.0 genes in this pathway |
| SOTIRIOU BREAST CANCER GRADE 1 VS 3 UP | 151 | 84 | All SZGR 2.0 genes in this pathway |
| WILCOX RESPONSE TO PROGESTERONE UP | 152 | 90 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS DN | 310 | 188 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL DN | 244 | 153 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS UP | 290 | 177 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY UP | 430 | 232 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS TURQUOISE DN | 53 | 25 | All SZGR 2.0 genes in this pathway |
| ODONNELL TFRC TARGETS DN | 139 | 76 | All SZGR 2.0 genes in this pathway |
| ODONNELL TARGETS OF MYC AND TFRC DN | 45 | 25 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN | 459 | 276 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA DN | 146 | 94 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML QUIESCENT VS NORMAL QUIESCENT UP | 87 | 45 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML DIVIDING VS NORMAL QUIESCENT UP | 181 | 101 | All SZGR 2.0 genes in this pathway |
| GRAHAM NORMAL QUIESCENT VS NORMAL DIVIDING DN | 87 | 49 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS UP | 293 | 179 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN DN | 172 | 112 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| TANG SENESCENCE TP53 TARGETS DN | 57 | 33 | All SZGR 2.0 genes in this pathway |
| JOHANSSON GLIOMAGENESIS BY PDGFB UP | 58 | 44 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 6 7WK DN | 79 | 54 | All SZGR 2.0 genes in this pathway |
| KONG E2F3 TARGETS | 97 | 58 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS BASAL | 330 | 217 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER BASAL VS LULMINAL | 330 | 215 | All SZGR 2.0 genes in this pathway |
| ROSTY CERVICAL CANCER PROLIFERATION CLUSTER | 140 | 73 | All SZGR 2.0 genes in this pathway |
| WAESCH ANAPHASE PROMOTING COMPLEX | 12 | 7 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| KORKOLA TERATOMA | 39 | 25 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM1 | 229 | 137 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| PUJANA XPRSS INT NETWORK | 168 | 103 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA CENTERED NETWORK | 117 | 72 | All SZGR 2.0 genes in this pathway |
| WILLIAMS ESR1 TARGETS UP | 26 | 16 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| SCIAN CELL CYCLE TARGETS OF TP53 AND TP73 DN | 22 | 14 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES 1 | 379 | 235 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| MORI PRE BI LYMPHOCYTE UP | 80 | 54 | All SZGR 2.0 genes in this pathway |
| MORI LARGE PRE BII LYMPHOCYTE UP | 86 | 49 | All SZGR 2.0 genes in this pathway |
| MORI IMMATURE B LYMPHOCYTE DN | 90 | 55 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO DN | 200 | 112 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB TARGETS | 74 | 41 | All SZGR 2.0 genes in this pathway |
| SHEPARD CRUSH AND BURN MUTANT DN | 185 | 111 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST DN | 309 | 206 | All SZGR 2.0 genes in this pathway |
| LE EGR2 TARGETS UP | 108 | 75 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 1 DN | 163 | 115 | All SZGR 2.0 genes in this pathway |
| ALCALAY AML BY NPM1 LOCALIZATION DN | 184 | 132 | All SZGR 2.0 genes in this pathway |
| HADDAD T LYMPHOCYTE AND NK PROGENITOR DN | 63 | 41 | All SZGR 2.0 genes in this pathway |
| LEE AGING NEOCORTEX DN | 80 | 49 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS BY HNE AND H2O2 | 39 | 29 | All SZGR 2.0 genes in this pathway |
| PAL PRMT5 TARGETS UP | 203 | 135 | All SZGR 2.0 genes in this pathway |
| KANG DOXORUBICIN RESISTANCE UP | 54 | 33 | All SZGR 2.0 genes in this pathway |
| RHODES UNDIFFERENTIATED CANCER | 69 | 44 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 3 | 101 | 64 | All SZGR 2.0 genes in this pathway |
| DELLA RESPONSE TO TSA AND BUTYRATE | 21 | 17 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS PEAK AT 24HR | 43 | 30 | All SZGR 2.0 genes in this pathway |
| LY AGING OLD DN | 56 | 35 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C7 | 68 | 44 | All SZGR 2.0 genes in this pathway |
| SIMBULAN PARP1 TARGETS DN | 17 | 10 | All SZGR 2.0 genes in this pathway |
| LY AGING MIDDLE DN | 16 | 10 | All SZGR 2.0 genes in this pathway |
| HU GENOTOXIC DAMAGE 4HR | 35 | 28 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P6 | 91 | 44 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| HELLER SILENCED BY METHYLATION UP | 282 | 183 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS UP | 317 | 208 | All SZGR 2.0 genes in this pathway |
| WONG PROTEASOME GENE MODULE | 49 | 35 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL A DN | 18 | 7 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| KUMAMOTO RESPONSE TO NUTLIN 3A DN | 10 | 5 | All SZGR 2.0 genes in this pathway |
| ZHENG GLIOBLASTOMA PLASTICITY UP | 250 | 168 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| BHATI G2M ARREST BY 2METHOXYESTRADIOL UP | 125 | 68 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| FRASOR RESPONSE TO SERM OR FULVESTRANT DN | 50 | 29 | All SZGR 2.0 genes in this pathway |
| GOLDRATH ANTIGEN RESPONSE | 346 | 192 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER ESR1 DN | 240 | 153 | All SZGR 2.0 genes in this pathway |
| HOFFMANN LARGE TO SMALL PRE BII LYMPHOCYTE UP | 163 | 102 | All SZGR 2.0 genes in this pathway |
| HOFFMANN SMALL PRE BII TO IMMATURE B LYMPHOCYTE DN | 50 | 33 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6 | 456 | 285 | All SZGR 2.0 genes in this pathway |
| COLINA TARGETS OF 4EBP1 AND 4EBP2 | 356 | 214 | All SZGR 2.0 genes in this pathway |
| ZHAN EARLY DIFFERENTIATION GENES DN | 42 | 29 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| CROONQUIST IL6 DEPRIVATION DN | 98 | 69 | All SZGR 2.0 genes in this pathway |
| CROONQUIST NRAS SIGNALING DN | 72 | 47 | All SZGR 2.0 genes in this pathway |
| CROONQUIST NRAS VS STROMAL STIMULATION DN | 99 | 65 | All SZGR 2.0 genes in this pathway |
| ISHIDA E2F TARGETS | 53 | 27 | All SZGR 2.0 genes in this pathway |
| POOLA INVASIVE BREAST CANCER UP | 288 | 168 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS PROLIFERATION UP | 178 | 108 | All SZGR 2.0 genes in this pathway |
| PUJANA BREAST CANCER WITH BRCA1 MUTATED UP | 56 | 27 | All SZGR 2.0 genes in this pathway |
| KOBAYASHI EGFR SIGNALING 24HR DN | 251 | 151 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S1 | 237 | 159 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING DN | 225 | 124 | All SZGR 2.0 genes in this pathway |
| WONG EMBRYONIC STEM CELL CORE | 335 | 193 | All SZGR 2.0 genes in this pathway |
| NAKAYAMA SOFT TISSUE TUMORS PCA2 UP | 87 | 50 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE LITERATURE | 44 | 25 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G2 M | 216 | 124 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS GROWING | 243 | 155 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
| SERVITJA LIVER HNF1A TARGETS UP | 135 | 96 | All SZGR 2.0 genes in this pathway |
| YANG BCL3 TARGETS UP | 364 | 236 | All SZGR 2.0 genes in this pathway |
| LE NEURONAL DIFFERENTIATION DN | 19 | 16 | All SZGR 2.0 genes in this pathway |
| FOSTER KDM1A TARGETS DN | 211 | 119 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL UP | 289 | 184 | All SZGR 2.0 genes in this pathway |
| SMIRNOV RESPONSE TO IR 6HR DN | 114 | 69 | All SZGR 2.0 genes in this pathway |
| ZHOU CELL CYCLE GENES IN IR RESPONSE 24HR | 128 | 73 | All SZGR 2.0 genes in this pathway |