Gene Page: CDKN1A
Summary ?
| GeneID | 1026 |
| Symbol | CDKN1A |
| Synonyms | CAP20|CDKN1|CIP1|MDA-6|P21|SDI1|WAF1|p21CIP1 |
| Description | cyclin-dependent kinase inhibitor 1A |
| Reference | MIM:116899|HGNC:HGNC:1784|Ensembl:ENSG00000124762|HPRD:00298|Vega:OTTHUMG00000014603 |
| Gene type | protein-coding |
| Map location | 6p21.2 |
| Pascal p-value | 0.948 |
| Sherlock p-value | 0.393 |
| Fetal beta | -1.715 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.033 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.04433 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.1164 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg04214946 | 6 | 36651933 | CDKN1A | 4.312E-4 | 0.417 | 0.045 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs9985704 | chr4 | 7050376 | CDKN1A | 1026 | 0.19 | trans | ||
| rs317714 | chr7 | 29115553 | CDKN1A | 1026 | 0.01 | trans | ||
| rs4921994 | chr8 | 18954801 | CDKN1A | 1026 | 0.11 | trans | ||
| rs11019015 | chr11 | 90157517 | CDKN1A | 1026 | 0.13 | trans | ||
| rs2993290 | chr13 | 113647189 | CDKN1A | 1026 | 0.18 | trans |
Section II. Transcriptome annotation
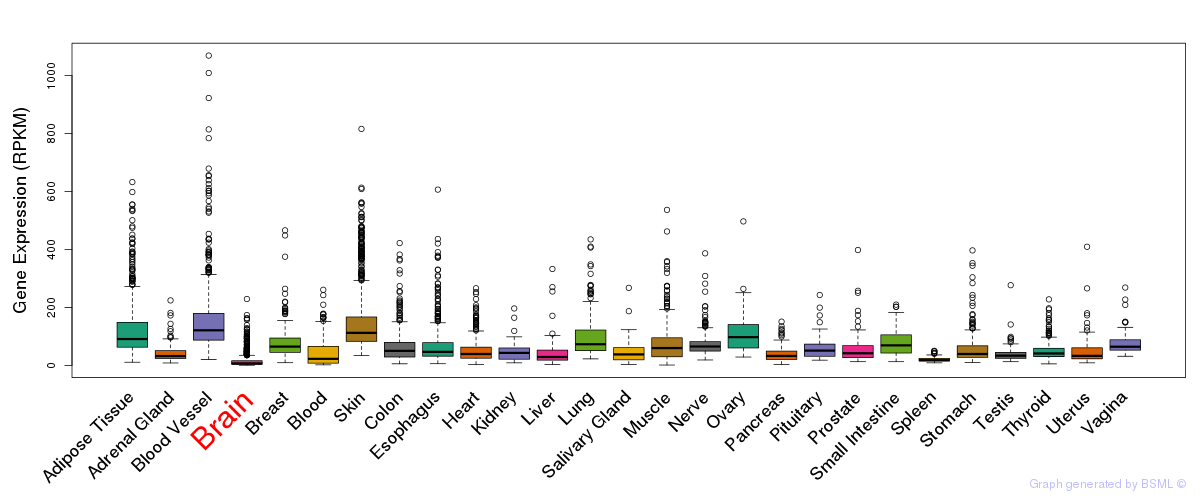
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ANKRD13D | 0.88 | 0.85 |
| SYT3 | 0.86 | 0.83 |
| MGAT4B | 0.85 | 0.86 |
| ACOT7 | 0.85 | 0.83 |
| PIP5KL1 | 0.83 | 0.78 |
| RTN2 | 0.83 | 0.85 |
| AC011479.2 | 0.83 | 0.81 |
| FKBP1B | 0.82 | 0.81 |
| CRIP2 | 0.82 | 0.86 |
| AC024575.2 | 0.82 | 0.86 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.8 | -0.62 | -0.62 |
| AF347015.33 | -0.62 | -0.62 |
| AF347015.27 | -0.60 | -0.62 |
| AF347015.26 | -0.60 | -0.63 |
| MT-CO2 | -0.60 | -0.58 |
| AF347015.15 | -0.59 | -0.59 |
| MT-CYB | -0.58 | -0.58 |
| MT-ATP8 | -0.58 | -0.64 |
| AF347015.2 | -0.57 | -0.57 |
| AF347015.21 | -0.57 | -0.58 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004861 | cyclin-dependent protein kinase inhibitor activity | EXP | 10323868 | |
| GO:0004861 | cyclin-dependent protein kinase inhibitor activity | IEA | - | |
| GO:0004861 | cyclin-dependent protein kinase inhibitor activity | TAS | 8242751 | |
| GO:0004860 | protein kinase inhibitor activity | IEA | - | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0016301 | kinase activity | IEA | - | |
| GO:0030332 | cyclin binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000082 | G1/S transition of mitotic cell cycle | IDA | 10208428 | |
| GO:0000086 | G2/M transition of mitotic cell cycle | IMP | 17553787 | |
| GO:0007050 | cell cycle arrest | IEA | - | |
| GO:0008285 | negative regulation of cell proliferation | IEA | - | |
| GO:0008629 | induction of apoptosis by intracellular signals | TAS | 9660939 | |
| GO:0009411 | response to UV | IEA | - | |
| GO:0006974 | response to DNA damage stimulus | IEA | - | |
| GO:0043066 | negative regulation of apoptosis | IEA | - | |
| GO:0042326 | negative regulation of phosphorylation | IDA | 10208428 | |
| GO:0043071 | positive regulation of non-apoptotic programmed cell death | IEA | - | |
| GO:0030890 | positive regulation of B cell proliferation | IEA | - | |
| GO:0030308 | negative regulation of cell growth | IDA | 10208428 | |
| GO:0031668 | cellular response to extracellular stimulus | IMP | 17553787 | |
| GO:0045736 | negative regulation of cyclin-dependent protein kinase activity | IEA | - | |
| GO:0048146 | positive regulation of fibroblast proliferation | IMP | 17420273 | |
| GO:0051726 | regulation of cell cycle | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000307 | cyclin-dependent protein kinase holoenzyme complex | IEA | - | |
| GO:0005829 | cytosol | EXP | 9106657 |9632134 | |
| GO:0005829 | cytosol | IEA | - | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005654 | nucleoplasm | EXP | 9190208 |9632134 |9840943 |10323868 |11231585 |11931757 |16782892 | |
| GO:0005737 | cytoplasm | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| APEX1 | APE | APE-1 | APE1 | APEN | APEX | APX | HAP1 | REF-1 | REF1 | APEX nuclease (multifunctional DNA repair enzyme) 1 | Ref1 interacts weakly with p21 promoter. | BIND | 15674341 |
| BCCIP | TOK-1 | TOK1 | BRCA2 and CDKN1A interacting protein | - | HPRD,BioGRID | 10878006 |14726710 |
| C7orf20 | CEE | CGI-20 | chromosome 7 open reading frame 20 | Two-hybrid | BioGRID | 16169070 |
| CCDC85B | DIPA | coiled-coil domain containing 85B | Two-hybrid | BioGRID | 16189514 |
| CCNA1 | - | cyclin A1 | cyclin A1 interacts with p21. | BIND | 10022926 |
| CCNA2 | CCN1 | CCNA | cyclin A2 | p21 interacts with cyclin A. | BIND | 9632134 |
| CCNA2 | CCN1 | CCNA | cyclin A2 | cyclin A interacts with p21. This interaction was modeled on a demonstrated interaction between monkey cyclin A and monkey p21. | BIND | 16009130 |
| CCNA2 | CCN1 | CCNA | cyclin A2 | CDKN1A (p21) interacts with CCNA2 (cyclin A). | BIND | 8662825 |
| CCNA2 | CCN1 | CCNA | cyclin A2 | Affinity Capture-Western | BioGRID | 12947099 |
| CCNB1 | CCNB | cyclin B1 | CDKN1A (p21) interacts with CCNB1 (cyclin B1). | BIND | 8662825 |
| CCND1 | BCL1 | D11S287E | PRAD1 | U21B31 | cyclin D1 | - | HPRD,BioGRID | 7478582 |8657154 |
| CCND1 | BCL1 | D11S287E | PRAD1 | U21B31 | cyclin D1 | p21 interacts with cyclin D. | BIND | 9632134 |
| CCND1 | BCL1 | D11S287E | PRAD1 | U21B31 | cyclin D1 | Cyclin D1 interacts with p21. | BIND | 8999999 |
| CCND1 | BCL1 | D11S287E | PRAD1 | U21B31 | cyclin D1 | CDKN1A (p21) interacts with CCND1 (cyclin D1). | BIND | 8662825 |
| CCND2 | KIAK0002 | MGC102758 | cyclin D2 | Two-hybrid | BioGRID | 16189514 |
| CCND3 | - | cyclin D3 | Two-hybrid | BioGRID | 16189514 |
| CCNE1 | CCNE | cyclin E1 | p21 interacts with cyclin E. | BIND | 9632134 |
| CCNE1 | CCNE | cyclin E1 | Affinity Capture-Western | BioGRID | 12839982 |
| CCNE1 | CCNE | cyclin E1 | CDKN1A (p21) interacts with CCNE1 (cyclin E). | BIND | 8662825 |
| CDC2 | CDC28A | CDK1 | DKFZp686L20222 | MGC111195 | cell division cycle 2, G1 to S and G2 to M | p21 interacts with CDC2. This interaction was modeled on a demonstrated interaction between human p21 and hamster CDC2. | BIND | 9467962 |
| CDC7 | CDC7L1 | HsCDC7 | Hsk1 | MGC117361 | MGC126237 | MGC126238 | huCDC7 | cell division cycle 7 homolog (S. cerevisiae) | - | HPRD | 15232106 |
| CDK2 | p33(CDK2) | cyclin-dependent kinase 2 | p21WAF1 interacts with an unspecified isoform of Cdk2. This interaction was modelled on a demonstrated interaction between human p21WAF1 and Cdk2 from an unspecified species. | BIND | 7780738 |
| CDK2 | p33(CDK2) | cyclin-dependent kinase 2 | p21 interacts with an unspecified isoform of Cdk2. | BIND | 11302688 |
| CDK2 | p33(CDK2) | cyclin-dependent kinase 2 | p21 interacts with CDK2. This interaction was modeled on a demonstrated interaction between human p21 and hamster CDK2. | BIND | 9467962 |
| CDK2 | p33(CDK2) | cyclin-dependent kinase 2 | p21 interacts with Cdk2. | BIND | 9546435 |
| CDK2 | p33(CDK2) | cyclin-dependent kinase 2 | - | HPRD,BioGRID | 12839982 |
| CDK4 | CMM3 | MGC14458 | PSK-J3 | cyclin-dependent kinase 4 | Cdk4 interacts with p21. | BIND | 8999999 |
| CDKN1B | CDKN4 | KIP1 | MEN1B | MEN4 | P27KIP1 | cyclin-dependent kinase inhibitor 1B (p27, Kip1) | - | HPRD | 9858585 |
| CHEK1 | CHK1 | CHK1 checkpoint homolog (S. pombe) | Affinity Capture-Western | BioGRID | 11896572 |
| CIZ1 | LSFR1 | NP94 | ZNF356 | CDKN1A interacting zinc finger protein 1 | - | HPRD,BioGRID | 10529385 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | CREBBP (CBP) interacts with the p21 promoter. | BIND | 15710329 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | CBP interacts with p21. | BIND | 10764767 |
| CSNK2A1 | CK2A1 | CKII | casein kinase 2, alpha 1 polypeptide | - | HPRD | 11255227 |
| CSNK2B | CK2B | CK2N | CSK2B | G5A | MGC138222 | MGC138224 | casein kinase 2, beta polypeptide | - | HPRD,BioGRID | 10679299 |
| DDX5 | DKFZp686J01190 | G17P1 | HLR1 | HUMP68 | p68 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 | p68 interacts with the p21 promoter. | BIND | 15660129 |
| FOXO1 | FKH1 | FKHR | FOXO1A | forkhead box O1 | FoxO1 binds the distal region of the p21Cip1 promoter. This interaction was modelled on a demonstrated interaction between FoxO1 from an unspecified species and p21Cip1 promoter from an unspecified species. | BIND | 15084259 |
| FOXO3 | AF6q21 | DKFZp781A0677 | FKHRL1 | FKHRL1P2 | FOXO2 | FOXO3A | MGC12739 | MGC31925 | forkhead box O3 | FoxO3 binds the distal region of the p21Cip1 promoter. | BIND | 15084259 |
| FOXO4 | AFX | AFX1 | MGC120490 | MLLT7 | forkhead box O4 | FoxO4 binds the distal region of the p21Cip1 promoter. This interaction was modelled on a demonstrated interaction between FoxO4 from an unspecified species and p21Cip1 promoter from an unspecified species. | BIND | 15084259 |
| GADD45A | DDIT1 | GADD45 | growth arrest and DNA-damage-inducible, alpha | - | HPRD | 7478594 |11498536 |
| GADD45A | DDIT1 | GADD45 | growth arrest and DNA-damage-inducible, alpha | Affinity Capture-Western Reconstituted Complex | BioGRID | 10912791 |10973963 |
| GADD45B | DKFZp566B133 | GADD45BETA | MYD118 | growth arrest and DNA-damage-inducible, beta | - | HPRD | 8700517 |
| GADD45G | CR6 | DDIT2 | GADD45gamma | GRP17 | growth arrest and DNA-damage-inducible, gamma | - | HPRD,BioGRID | 10455148 |
| GNB2L1 | Gnb2-rs1 | H12.3 | HLC-7 | PIG21 | RACK1 | guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 | Two-hybrid | BioGRID | 16169070 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | HDAC1 interacts with the p21 promoter. | BIND | 15710329 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | HDAC1 interacts with the CDKN1A (p21) promoter. | BIND | 15674334 |
| HDAC11 | FLJ22237 | histone deacetylase 11 | Two-hybrid | BioGRID | 16169070 |
| HERC5 | CEB1 | CEBP1 | hect domain and RLD 5 | Reconstituted Complex | BioGRID | 10581175 |
| HIF1A | HIF-1alpha | HIF1 | HIF1-ALPHA | MOP1 | PASD8 | hypoxia-inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) | HIF-1alpha interacts with CDKN1A promoter. | BIND | 15780936 |
| HIST4H4 | H4/p | MGC24116 | histone cluster 4, H4 | HIST4H4 (H4) interacts with the CDKN1A promoter. | BIND | 15765097 |
| HIST4H4 | H4/p | MGC24116 | histone cluster 4, H4 | HIST4H4 (H4) interacts with the p21 promoter. | BIND | 15710329 |
| MAP3K5 | ASK1 | MAPKKK5 | MEKK5 | mitogen-activated protein kinase kinase kinase 5 | Affinity Capture-Western | BioGRID | 12820963 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | Mdm2 interacts with the p21 promoter. | BIND | 15546622 |
| MED1 | CRSP1 | CRSP200 | DRIP205 | DRIP230 | MGC71488 | PBP | PPARBP | PPARGBP | RB18A | TRAP220 | TRIP2 | mediator complex subunit 1 | MED1 interacts with p21 promoter. | BIND | 15989967 |
| MED21 | SRB7 | SURB7 | mediator complex subunit 21 | MED21 interacts with p21 promoter. | BIND | 15989967 |
| MITF | MI | WS2A | bHLHe32 | microphthalmia-associated transcription factor | An unspecified isoform of Mitf interacts with the p21 promoter. | BIND | 15716956 |
| MUC1 | CD227 | EMA | H23AG | MAM6 | PEM | PEMT | PUM | mucin 1, cell surface associated | MUC1 C-ter interacts with the p21 promoter. | BIND | 15710329 |
| MYC | bHLHe39 | c-Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | Myc interacts with CDKN1A promoter. | BIND | 15780936 |
| MYC | bHLHe39 | c-Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | c-Myc binds the proximal region of the p21Cip1 promoter. | BIND | 15084259 |
| MYC | bHLHe39 | c-Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | c-Myc interacts with p21WAF1 promoter. | BIND | 15856024 |
| PARP1 | ADPRT | ADPRT1 | PARP | PARP-1 | PPOL | pADPRT-1 | poly (ADP-ribose) polymerase 1 | - | HPRD,BioGRID | 12930846 |
| PCNA | MGC8367 | proliferating cell nuclear antigen | CDKN1A (p21) interacts with PCNA. | BIND | 8662825 |
| PCNA | MGC8367 | proliferating cell nuclear antigen | p21WAF1 interacts with PCNA. | BIND | 7780738 |
| PCNA | MGC8367 | proliferating cell nuclear antigen | PCNA interacts with p21. | BIND | 9705499 |
| PCNA | MGC8367 | proliferating cell nuclear antigen | PCNA interacts with p21. | BIND | 9178907 |
| PCNA | MGC8367 | proliferating cell nuclear antigen | p21 interacts with PCNA. | BIND | 11302688 |
| PCNA | MGC8367 | proliferating cell nuclear antigen | PCNA interacts with p21. | BIND | 8861969 |
| PCNA | MGC8367 | proliferating cell nuclear antigen | p21 interacts with PCNA. | BIND | 9632134 |
| PCNA | MGC8367 | proliferating cell nuclear antigen | p21 interacts with PCNA. This interaction was modeled on a demonstrated interaction between human p21 and PCNA from an unspecified species. | BIND | 9546435 |
| PCNA | MGC8367 | proliferating cell nuclear antigen | - | HPRD,BioGRID | 8861913 |
| PCNA | MGC8367 | proliferating cell nuclear antigen | p21WAF1 interacts with PCNA. This interaction was modeled on a demonstrated interaction between human p21WAF1 and Pisum sativum v. Onward PCNA. | BIND | 8647134 |
| PCNA | MGC8367 | proliferating cell nuclear antigen | p21 interacts with PCNA. This interaction was modeled on a demonstrated interaction between human p21 and hamster PCNA. | BIND | 9467962 |
| PIAS2 | MGC102682 | MIZ1 | PIASX | PIASX-ALPHA | PIASX-BETA | SIZ2 | ZMIZ4 | miz | protein inhibitor of activated STAT, 2 | Miz1 interacts with p-P21CIP1. | BIND | 15580267 |
| PIAS2 | MGC102682 | MIZ1 | PIASX | PIASX-ALPHA | PIASX-BETA | SIZ2 | ZMIZ4 | miz | protein inhibitor of activated STAT, 2 | Miz-1 interacts with CDKN1A promoter. | BIND | 15780936 |
| PIM1 | PIM | pim-1 oncogene | - | HPRD,BioGRID | 12431783 |
| POLD2 | - | polymerase (DNA directed), delta 2, regulatory subunit 50kDa | - | HPRD | 12522211 |
| POLD2 | - | polymerase (DNA directed), delta 2, regulatory subunit 50kDa | Waf1 interacts with p50 subunit of DNA polymerase delta. | BIND | 12522211 |
| PSMA3 | HC8 | MGC12306 | MGC32631 | PSC3 | proteasome (prosome, macropain) subunit, alpha type, 3 | - | HPRD,BioGRID | 11350925 |
| RAB1A | DKFZp564B163 | RAB1 | YPT1 | RAB1A, member RAS oncogene family | Two-hybrid | BioGRID | 16169070 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | Rb interacts with the p21 promoter. | BIND | 15716956 |
| RNF144B | IBRDC2 | KIAA0161 | MGC71786 | bA528A10.3 | p53RFP | ring finger 144B | - | HPRD | 12853982 |
| SET | 2PP2A | I2PP2A | IGAAD | IPP2A2 | PHAPII | TAF-I | TAF-IBETA | SET nuclear oncogene | - | HPRD,BioGRID | 12407107 |
| SMAD4 | DPC4 | JIP | MADH4 | SMAD family member 4 | Smad4 binds the distal region of the p21Cip1 promoter. | BIND | 15084259 |
| SMARCA4 | BAF190 | BRG1 | FLJ39786 | SNF2 | SNF2-BETA | SNF2L4 | SNF2LB | SWI2 | hSNF2b | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 | BRG1 interacts with p21 promoter. | BIND | 15780937 |
| SP1 | - | Sp1 transcription factor | SP1 interacts with the CDKN1A (p21) promoter. | BIND | 15674334 |
| SP1 | - | Sp1 transcription factor | Sp1 interacts with CDKN1A promoter. | BIND | 15780936 |
| STAT3 | APRF | FLJ20882 | HIES | MGC16063 | signal transducer and activator of transcription 3 (acute-phase response factor) | - | HPRD,BioGRID | 10764767 |
| TEX11 | TGC1 | TSGA3 | testis expressed 11 | Two-hybrid | BioGRID | 16189514 |
| TK1 | TK2 | thymidine kinase 1, soluble | - | HPRD,BioGRID | 11389691 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | p53 interacts with p21 promoter. | BIND | 16009130 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | TP53 (p53) interacts with the CDKN1A (p21) promoter. | BIND | 15674334 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | p53 interacts with the p21 promoter. | BIND | 15660129 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | Affinity Capture-Western | BioGRID | 11896572 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | p53 interacts with p21 promoter. | BIND | 15989967 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | p53 interacts with the p21 promoter. | BIND | 15710329 |
| TP73 | P73 | tumor protein p73 | p73-beta interacts with p21 promoter. This interaction was modeled on a demonstrated interaction between p73-beta from an unspecified species and human p21 promoter. | BIND | 15893728 |
| TP73 | P73 | tumor protein p73 | p73-alpha interacts with p21 promoter. | BIND | 15893728 |
| TSG101 | TSG10 | VPS23 | tumor susceptibility gene 101 | - | HPRD,BioGRID | 11943869 |
| ZBTB17 | MIZ-1 | ZNF151 | ZNF60 | pHZ-67 | zinc finger and BTB domain containing 17 | Miz1 interacts with p21WAF1 promoter. | BIND | 15856024 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ERBB SIGNALING PATHWAY | 87 | 71 | All SZGR 2.0 genes in this pathway |
| KEGG CELL CYCLE | 128 | 84 | All SZGR 2.0 genes in this pathway |
| KEGG P53 SIGNALING PATHWAY | 69 | 45 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG GLIOMA | 65 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG PROSTATE CANCER | 89 | 75 | All SZGR 2.0 genes in this pathway |
| KEGG MELANOMA | 71 | 57 | All SZGR 2.0 genes in this pathway |
| KEGG BLADDER CANCER | 42 | 33 | All SZGR 2.0 genes in this pathway |
| KEGG CHRONIC MYELOID LEUKEMIA | 73 | 59 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ATM PATHWAY | 20 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA G1 PATHWAY | 28 | 21 | All SZGR 2.0 genes in this pathway |
| BIOCARTA G2 PATHWAY | 24 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CELLCYCLE PATHWAY | 23 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CALCINEURIN PATHWAY | 21 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EPONFKB PATHWAY | 11 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA P53HYPOXIA PATHWAY | 23 | 21 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RACCYCD PATHWAY | 26 | 23 | All SZGR 2.0 genes in this pathway |
| BIOCARTA P53 PATHWAY | 16 | 13 | All SZGR 2.0 genes in this pathway |
| SA G1 AND S PHASES | 15 | 10 | All SZGR 2.0 genes in this pathway |
| SA G2 AND M PHASES | 8 | 7 | All SZGR 2.0 genes in this pathway |
| SA TRKA RECEPTOR | 17 | 15 | All SZGR 2.0 genes in this pathway |
| PID SMAD2 3NUCLEAR PATHWAY | 82 | 63 | All SZGR 2.0 genes in this pathway |
| PID PRL SIGNALING EVENTS PATHWAY | 23 | 17 | All SZGR 2.0 genes in this pathway |
| PID NOTCH PATHWAY | 59 | 49 | All SZGR 2.0 genes in this pathway |
| PID P73PATHWAY | 79 | 59 | All SZGR 2.0 genes in this pathway |
| PID HDAC CLASSIII PATHWAY | 25 | 20 | All SZGR 2.0 genes in this pathway |
| PID E2F PATHWAY | 74 | 48 | All SZGR 2.0 genes in this pathway |
| PID ANGIOPOIETIN RECEPTOR PATHWAY | 50 | 41 | All SZGR 2.0 genes in this pathway |
| PID REG GR PATHWAY | 82 | 60 | All SZGR 2.0 genes in this pathway |
| PID P53 DOWNSTREAM PATHWAY | 137 | 94 | All SZGR 2.0 genes in this pathway |
| PID CMYB PATHWAY | 84 | 61 | All SZGR 2.0 genes in this pathway |
| PID PI3KCI AKT PATHWAY | 35 | 30 | All SZGR 2.0 genes in this pathway |
| PID MYC REPRESS PATHWAY | 63 | 52 | All SZGR 2.0 genes in this pathway |
| PID TAP63 PATHWAY | 54 | 40 | All SZGR 2.0 genes in this pathway |
| PID RB 1PATHWAY | 65 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY SCF KIT | 78 | 59 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB4 | 90 | 67 | All SZGR 2.0 genes in this pathway |
| REACTOME ORC1 REMOVAL FROM CHROMATIN | 67 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB2 | 101 | 78 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY EGFR IN CANCER | 109 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K EVENTS IN ERBB4 SIGNALING | 38 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K EVENTS IN ERBB2 SIGNALING | 44 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | 97 | 66 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | 126 | 90 | All SZGR 2.0 genes in this pathway |
| REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | 137 | 105 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR IN DISEASE | 127 | 88 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K AKT ACTIVATION | 38 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | 12 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME GAB1 SIGNALOSOME | 38 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE CHECKPOINTS | 124 | 70 | All SZGR 2.0 genes in this pathway |
| REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | 65 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME G1 PHASE | 38 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | 57 | 35 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY PDGF | 122 | 93 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | 95 | 76 | All SZGR 2.0 genes in this pathway |
| REACTOME G1 S TRANSITION | 112 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS OF DNA | 92 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC G1 G1 S PHASES | 137 | 79 | All SZGR 2.0 genes in this pathway |
| REACTOME PI 3K CASCADE | 56 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNALING OF ACTIVATED FGFR | 100 | 74 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPLICATION | 192 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME PIP3 ACTIVATES AKT SIGNALING | 29 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME S PHASE | 109 | 66 | All SZGR 2.0 genes in this pathway |
| REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | 56 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR | 112 | 80 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| KIM RESPONSE TO TSA AND DECITABINE UP | 129 | 73 | All SZGR 2.0 genes in this pathway |
| CHEMNITZ RESPONSE TO PROSTAGLANDIN E2 DN | 391 | 222 | All SZGR 2.0 genes in this pathway |
| ZERBINI RESPONSE TO SULINDAC DN | 6 | 5 | All SZGR 2.0 genes in this pathway |
| ZHONG RESPONSE TO AZACITIDINE AND TSA UP | 183 | 119 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL UP | 276 | 187 | All SZGR 2.0 genes in this pathway |
| DOANE RESPONSE TO ANDROGEN DN | 241 | 146 | All SZGR 2.0 genes in this pathway |
| NEWMAN ERCC6 TARGETS DN | 39 | 24 | All SZGR 2.0 genes in this pathway |
| SMIRNOV CIRCULATING ENDOTHELIOCYTES IN CANCER UP | 158 | 103 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL UP | 233 | 161 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS BLACK UP | 35 | 22 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS DN | 240 | 171 | All SZGR 2.0 genes in this pathway |
| ODONNELL TFRC TARGETS UP | 456 | 228 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES CD4 UP | 64 | 46 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 48HR UP | 128 | 95 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 96HR UP | 117 | 84 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| LAU APOPTOSIS CDKN2A UP | 55 | 40 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION KRAS CDC25 UP | 58 | 39 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 CHRONIC LOF UP | 115 | 78 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN UP | 239 | 157 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN UP | 612 | 367 | All SZGR 2.0 genes in this pathway |
| CAIRO PML TARGETS BOUND BY MYC DN | 14 | 12 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| KERLEY RESPONSE TO CISPLATIN UP | 44 | 30 | All SZGR 2.0 genes in this pathway |
| MISSIAGLIA REGULATED BY METHYLATION UP | 126 | 78 | All SZGR 2.0 genes in this pathway |
| TANG SENESCENCE TP53 TARGETS UP | 33 | 20 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 3 4WK DN | 39 | 27 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL TGFB1 TARGETS UP | 169 | 127 | All SZGR 2.0 genes in this pathway |
| NUNODA RESPONSE TO DASATINIB IMATINIB DN | 13 | 10 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| SANCHEZ MDM2 TARGETS | 15 | 10 | All SZGR 2.0 genes in this pathway |
| HUMMERICH SKIN CANCER PROGRESSION UP | 88 | 58 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 2 | 127 | 92 | All SZGR 2.0 genes in this pathway |
| SCHAVOLT TARGETS OF TP53 AND TP63 | 16 | 12 | All SZGR 2.0 genes in this pathway |
| KOMMAGANI TP63 GAMMA TARGETS | 9 | 7 | All SZGR 2.0 genes in this pathway |
| HERNANDEZ ABERRANT MITOSIS BY DOCETACEL 4NM UP | 23 | 15 | All SZGR 2.0 genes in this pathway |
| AMUNDSON DNA DAMAGE RESPONSE TP53 | 16 | 9 | All SZGR 2.0 genes in this pathway |
| SEITZ NEOPLASTIC TRANSFORMATION BY 8P DELETION DN | 30 | 25 | All SZGR 2.0 genes in this pathway |
| MEINHOLD OVARIAN CANCER LOW GRADE UP | 19 | 15 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS NEUROEPITHELIUM UP | 176 | 111 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER UP | 227 | 137 | All SZGR 2.0 genes in this pathway |
| MANN RESPONSE TO AMIFOSTINE UP | 20 | 12 | All SZGR 2.0 genes in this pathway |
| MURATA VIRULENCE OF H PILORI | 24 | 16 | All SZGR 2.0 genes in this pathway |
| TSAI RESPONSE TO IONIZING RADIATION | 149 | 101 | All SZGR 2.0 genes in this pathway |
| KHETCHOUMIAN TRIM24 TARGETS UP | 47 | 38 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| SCIAN CELL CYCLE TARGETS OF TP53 AND TP73 UP | 9 | 7 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY DN | 382 | 224 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS MODERATELY DN | 110 | 64 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS DN | 261 | 155 | All SZGR 2.0 genes in this pathway |
| FRIDMAN SENESCENCE UP | 77 | 60 | All SZGR 2.0 genes in this pathway |
| FRIDMAN IMMORTALIZATION DN | 34 | 24 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION DN | 206 | 136 | All SZGR 2.0 genes in this pathway |
| HE PTEN TARGETS UP | 16 | 15 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 120 HELA | 69 | 47 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO IKK INHIBITOR AND TNF UP | 223 | 140 | All SZGR 2.0 genes in this pathway |
| MORI PRE BI LYMPHOCYTE UP | 80 | 54 | All SZGR 2.0 genes in this pathway |
| MORI LARGE PRE BII LYMPHOCYTE UP | 86 | 49 | All SZGR 2.0 genes in this pathway |
| MORI SMALL PRE BII LYMPHOCYTE DN | 76 | 52 | All SZGR 2.0 genes in this pathway |
| MORI IMMATURE B LYMPHOCYTE DN | 90 | 55 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| LE EGR2 TARGETS DN | 108 | 84 | All SZGR 2.0 genes in this pathway |
| PENG LEUCINE DEPRIVATION UP | 142 | 93 | All SZGR 2.0 genes in this pathway |
| KANNAN TP53 TARGETS UP | 58 | 40 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST DN | 309 | 206 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER CIPROFIBRATE UP | 60 | 42 | All SZGR 2.0 genes in this pathway |
| NING CHRONIC OBSTRUCTIVE PULMONARY DISEASE UP | 157 | 105 | All SZGR 2.0 genes in this pathway |
| COLLER MYC TARGETS DN | 9 | 5 | All SZGR 2.0 genes in this pathway |
| LEI MYB TARGETS | 318 | 215 | All SZGR 2.0 genes in this pathway |
| NEMETH INFLAMMATORY RESPONSE LPS UP | 88 | 64 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA UP | 64 | 46 | All SZGR 2.0 genes in this pathway |
| PETROVA ENDOTHELIUM LYMPHATIC VS BLOOD DN | 162 | 102 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS UP | 214 | 133 | All SZGR 2.0 genes in this pathway |
| VERNELL RETINOBLASTOMA PATHWAY UP | 70 | 47 | All SZGR 2.0 genes in this pathway |
| MA MYELOID DIFFERENTIATION UP | 39 | 29 | All SZGR 2.0 genes in this pathway |
| ABE VEGFA TARGETS 2HR | 34 | 21 | All SZGR 2.0 genes in this pathway |
| BRACHAT RESPONSE TO CISPLATIN | 22 | 11 | All SZGR 2.0 genes in this pathway |
| WHITESIDE CISPLATIN RESISTANCE UP | 11 | 7 | All SZGR 2.0 genes in this pathway |
| GENTILE UV LOW DOSE UP | 27 | 19 | All SZGR 2.0 genes in this pathway |
| ONGUSAHA TP53 TARGETS | 38 | 23 | All SZGR 2.0 genes in this pathway |
| HARRIS HYPOXIA | 81 | 64 | All SZGR 2.0 genes in this pathway |
| HENDRICKS SMARCA4 TARGETS UP | 56 | 35 | All SZGR 2.0 genes in this pathway |
| RAMALHO STEMNESS UP | 206 | 118 | All SZGR 2.0 genes in this pathway |
| MCDOWELL ACUTE LUNG INJURY UP | 45 | 29 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 2 | 50 | 34 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 24HR DN | 148 | 102 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE UP | 244 | 165 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC DN | 228 | 146 | All SZGR 2.0 genes in this pathway |
| MURAKAMI UV RESPONSE 6HR UP | 37 | 31 | All SZGR 2.0 genes in this pathway |
| SIMBULAN PARP1 TARGETS UP | 31 | 23 | All SZGR 2.0 genes in this pathway |
| KANG FLUOROURACIL RESISTANCE UP | 22 | 15 | All SZGR 2.0 genes in this pathway |
| DELASERNA MYOD TARGETS UP | 89 | 51 | All SZGR 2.0 genes in this pathway |
| INGA TP53 TARGETS | 17 | 13 | All SZGR 2.0 genes in this pathway |
| CHIBA RESPONSE TO TSA | 50 | 31 | All SZGR 2.0 genes in this pathway |
| CHEN LVAD SUPPORT OF FAILING HEART UP | 103 | 69 | All SZGR 2.0 genes in this pathway |
| BRACHAT RESPONSE TO METHOTREXATE UP | 26 | 16 | All SZGR 2.0 genes in this pathway |
| MACLACHLAN BRCA1 TARGETS UP | 21 | 16 | All SZGR 2.0 genes in this pathway |
| GAJATE RESPONSE TO TRABECTEDIN UP | 67 | 45 | All SZGR 2.0 genes in this pathway |
| DELLA RESPONSE TO TSA AND BUTYRATE | 21 | 17 | All SZGR 2.0 genes in this pathway |
| BRACHAT RESPONSE TO CAMPTOTHECIN UP | 28 | 18 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS UP | 198 | 132 | All SZGR 2.0 genes in this pathway |
| JACKSON DNMT1 TARGETS UP | 77 | 57 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C1 | 72 | 45 | All SZGR 2.0 genes in this pathway |
| SEMENZA HIF1 TARGETS | 36 | 32 | All SZGR 2.0 genes in this pathway |
| VARELA ZMPSTE24 TARGETS UP | 40 | 30 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1 | 419 | 273 | All SZGR 2.0 genes in this pathway |
| KRIGE AMINO ACID DEPRIVATION | 29 | 20 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN | 409 | 268 | All SZGR 2.0 genes in this pathway |
| KANG CISPLATIN RESISTANCE UP | 19 | 11 | All SZGR 2.0 genes in this pathway |
| HELLER SILENCED BY METHYLATION UP | 282 | 183 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS UP | 317 | 208 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| FUJII YBX1 TARGETS UP | 43 | 28 | All SZGR 2.0 genes in this pathway |
| OUYANG PROSTATE CANCER MARKERS | 19 | 16 | All SZGR 2.0 genes in this pathway |
| HUANG FOXA2 TARGETS DN | 36 | 21 | All SZGR 2.0 genes in this pathway |
| SAGIV CD24 TARGETS DN | 46 | 26 | All SZGR 2.0 genes in this pathway |
| CHUNG BLISTER CYTOTOXICITY UP | 134 | 84 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB UP | 245 | 159 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA METAGENE | 242 | 168 | All SZGR 2.0 genes in this pathway |
| ALONSO METASTASIS UP | 198 | 128 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C D UP | 139 | 95 | All SZGR 2.0 genes in this pathway |
| HELLEBREKERS SILENCED DURING TUMOR ANGIOGENESIS | 80 | 56 | All SZGR 2.0 genes in this pathway |
| REICHERT G1S REGULATORS AS PI3K TARGETS | 8 | 6 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP | 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF DN | 235 | 144 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 DN | 245 | 150 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE DN | 209 | 137 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| SWEET KRAS ONCOGENIC SIGNATURE | 89 | 56 | All SZGR 2.0 genes in this pathway |
| HINATA NFKB TARGETS KERATINOCYTE UP | 91 | 63 | All SZGR 2.0 genes in this pathway |
| HOFFMANN LARGE TO SMALL PRE BII LYMPHOCYTE UP | 163 | 102 | All SZGR 2.0 genes in this pathway |
| HOFMANN MYELODYSPLASTIC SYNDROM LOW RISK UP | 22 | 15 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR UP | 221 | 150 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR UP | 293 | 203 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS UP | 108 | 78 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN PARTIALLY REPROGRAMMED TO PLURIPOTENCY | 10 | 6 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME3 AND H3K27ME3 | 142 | 103 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE UP | 181 | 106 | All SZGR 2.0 genes in this pathway |
| ONO FOXP3 TARGETS DN | 42 | 23 | All SZGR 2.0 genes in this pathway |
| TSAI DNAJB4 TARGETS UP | 13 | 8 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN | 253 | 192 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K4ME3 AND H3K27ME3 | 210 | 148 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE LITERATURE | 44 | 25 | All SZGR 2.0 genes in this pathway |
| WU APOPTOSIS BY CDKN1A VIA TP53 | 55 | 31 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER UP | 307 | 182 | All SZGR 2.0 genes in this pathway |
| QI HYPOXIA | 140 | 96 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR DN | 505 | 328 | All SZGR 2.0 genes in this pathway |
| WAGSCHAL EHMT2 TARGETS UP | 13 | 7 | All SZGR 2.0 genes in this pathway |
| IVANOVSKA MIR106B TARGETS | 90 | 56 | All SZGR 2.0 genes in this pathway |
| ZHU SKIL TARGETS UP | 20 | 14 | All SZGR 2.0 genes in this pathway |
| KIM PTEN TARGETS UP | 18 | 9 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS DN | 315 | 215 | All SZGR 2.0 genes in this pathway |
| KASLER HDAC7 TARGETS 2 DN | 32 | 23 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 7 | 403 | 240 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| HUANG GATA2 TARGETS UP | 149 | 96 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE VIA P38 PARTIAL | 160 | 106 | All SZGR 2.0 genes in this pathway |
| FOSTER KDM1A TARGETS UP | 266 | 142 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |
| SMIRNOV RESPONSE TO IR 2HR UP | 53 | 33 | All SZGR 2.0 genes in this pathway |
| SMIRNOV RESPONSE TO IR 6HR UP | 166 | 97 | All SZGR 2.0 genes in this pathway |
| GHANDHI DIRECT IRRADIATION UP | 110 | 68 | All SZGR 2.0 genes in this pathway |
| WARTERS RESPONSE TO IR SKIN | 83 | 44 | All SZGR 2.0 genes in this pathway |
| WARTERS IR RESPONSE 5GY | 47 | 23 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-17-5p/20/93.mr/106/519.d | 468 | 474 | m8 | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-208 | 86 | 92 | 1A | hsa-miR-208 | AUAAGACGAGCAAAAAGCUUGU |
| miR-22 | 951 | 957 | m8 | hsa-miR-22brain | AAGCUGCCAGUUGAAGAACUGU |
| miR-499 | 86 | 92 | 1A | hsa-miR-499 | UUAAGACUUGCAGUGAUGUUUAA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.