| PID ERBB4 PATHWAY
| 38 | 32 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 INTERNALIZATION PATHWAY
| 41 | 35 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY EGFR IN CANCER
| 109 | 80 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN
| 634 | 384 | All SZGR 2.0 genes in this pathway |
| LIU PROSTATE CANCER UP
| 96 | 57 | All SZGR 2.0 genes in this pathway |
| DOANE BREAST CANCER CLASSES UP
| 72 | 38 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY DN
| 362 | 238 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN
| 805 | 505 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN
| 537 | 339 | All SZGR 2.0 genes in this pathway |
| PROVENZANI METASTASIS UP
| 194 | 112 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP
| 552 | 347 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN UP
| 239 | 157 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS C UP
| 170 | 114 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2B
| 392 | 251 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN
| 712 | 443 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN
| 855 | 609 | All SZGR 2.0 genes in this pathway |
| HATADA METHYLATED IN LUNG CANCER UP
| 390 | 236 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES 1
| 379 | 235 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215
| 893 | 528 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 UP
| 256 | 159 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS UP
| 266 | 171 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS UP
| 126 | 84 | All SZGR 2.0 genes in this pathway |
| HADDAD T LYMPHOCYTE AND NK PROGENITOR UP
| 78 | 56 | All SZGR 2.0 genes in this pathway |
| ALCALAY AML BY NPM1 LOCALIZATION UP
| 140 | 83 | All SZGR 2.0 genes in this pathway |
| STOSSI RESPONSE TO ESTRADIOL
| 50 | 35 | All SZGR 2.0 genes in this pathway |
| HADDAD B LYMPHOCYTE PROGENITOR
| 293 | 193 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS DN
| 234 | 137 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN UP
| 262 | 186 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN
| 434 | 302 | All SZGR 2.0 genes in this pathway |
| BILD E2F3 ONCOGENIC SIGNATURE
| 246 | 153 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE UP
| 393 | 244 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 4
| 307 | 185 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER T4
| 94 | 69 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3
| 491 | 310 | All SZGR 2.0 genes in this pathway |
| ZHENG FOXP3 TARGETS IN THYMUS UP
| 196 | 137 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE DN
| 258 | 160 | All SZGR 2.0 genes in this pathway |
| MASSARWEH RESPONSE TO ESTRADIOL
| 61 | 47 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP
| 863 | 514 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN
| 701 | 446 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP
| 374 | 247 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C UP
| 47 | 29 | All SZGR 2.0 genes in this pathway |
| WU SILENCED BY METHYLATION IN BLADDER CANCER
| 55 | 42 | All SZGR 2.0 genes in this pathway |
| RUIZ TNC TARGETS UP
| 153 | 107 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER ESR1 UP
| 167 | 99 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK
| 1210 | 725 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED
| 351 | 238 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT
| 572 | 352 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS DN
| 414 | 237 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 6HR UP
| 229 | 149 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR UP
| 324 | 193 | All SZGR 2.0 genes in this pathway |
| PANGAS TUMOR SUPPRESSION BY SMAD1 AND SMAD5 UP
| 134 | 85 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS NOT VIA AKT1 UP
| 211 | 131 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS VIA AKT1 UP
| 281 | 183 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE EARLY LATE
| 317 | 190 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP
| 745 | 475 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN
| 553 | 343 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN
| 1080 | 713 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 7
| 403 | 240 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38
| 337 | 236 | All SZGR 2.0 genes in this pathway |
| VANOEVELEN MYOGENESIS SIN3A TARGETS
| 220 | 133 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY
| 1839 | 928 | All SZGR 2.0 genes in this pathway |
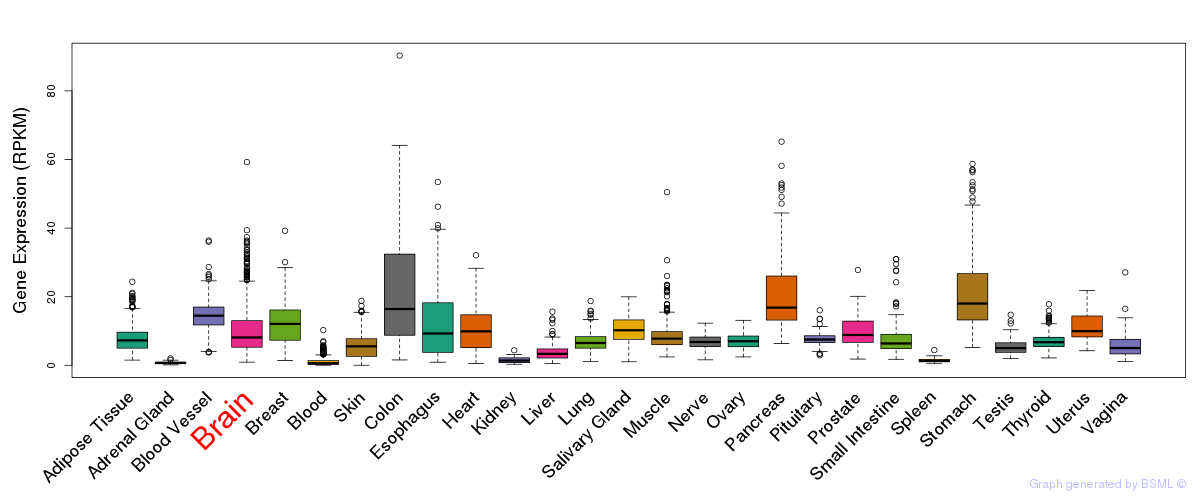
 Differentially methylated gene
Differentially methylated gene eQTL annotation
eQTL annotation