Gene Page: IFNG
Summary ?
| GeneID | 3458 |
| Symbol | IFNG |
| Synonyms | IFG|IFI |
| Description | interferon, gamma |
| Reference | MIM:147570|HGNC:HGNC:5438|Ensembl:ENSG00000111537|HPRD:00957|Vega:OTTHUMG00000169113 |
| Gene type | protein-coding |
| Map location | 12q14 |
| Pascal p-value | 0.719 |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16869851 | chr4 | 20690056 | IFNG | 3458 | 0.04 | trans | ||
| rs10994209 | chr10 | 61877705 | IFNG | 3458 | 0.15 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
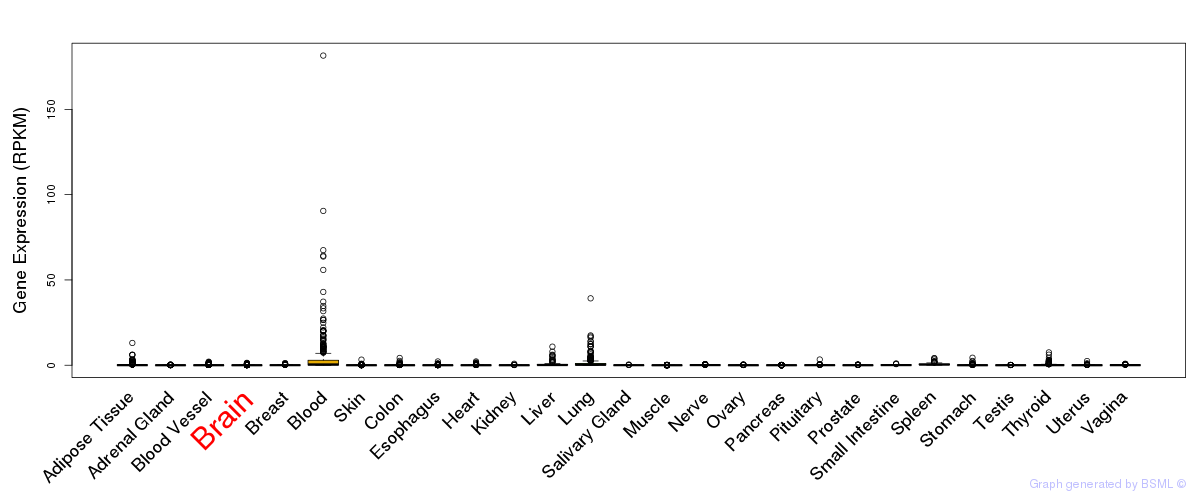
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005133 | interferon-gamma receptor binding | TAS | 10477596 | |
| GO:0005125 | cytokine activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0031642 | negative regulation of myelination | IEA | axon (GO term level: 15) | - |
| GO:0000060 | protein import into nucleus, translocation | IDA | 15604419 | |
| GO:0001558 | regulation of cell growth | IEA | - | |
| GO:0001781 | neutrophil apoptosis | IEA | - | |
| GO:0007166 | cell surface receptor linked signal transduction | TAS | 10477596 | |
| GO:0007050 | cell cycle arrest | IDA | 16942485 | |
| GO:0019882 | antigen processing and presentation | IEA | - | |
| GO:0048304 | positive regulation of isotype switching to IgG isotypes | IEA | - | |
| GO:0009615 | response to virus | IEA | - | |
| GO:0006928 | cell motion | TAS | 10477596 | |
| GO:0006925 | inflammatory cell apoptosis | IEA | - | |
| GO:0042508 | tyrosine phosphorylation of Stat1 protein | IDA | 15604419 | |
| GO:0030968 | endoplasmic reticulum unfolded protein response | IEA | - | |
| GO:0042742 | defense response to bacterium | IEA | - | |
| GO:0032735 | positive regulation of interleukin-12 production | IDA | 7605994 |8557999 | |
| GO:0034393 | positive regulation of smooth muscle cell apoptosis | IDA | 16942485 | |
| GO:0030593 | neutrophil chemotaxis | IEA | - | |
| GO:0045893 | positive regulation of transcription, DNA-dependent | IEA | - | |
| GO:0050718 | positive regulation of interleukin-1 beta secretion | IEA | - | |
| GO:0050776 | regulation of immune response | IEA | - | |
| GO:0045084 | positive regulation of interleukin-12 biosynthetic process | IEA | - | |
| GO:0045080 | positive regulation of chemokine biosynthetic process | IEA | - | |
| GO:0045348 | positive regulation of MHC class II biosynthetic process | IEA | - | |
| GO:0048662 | negative regulation of smooth muscle cell proliferation | IDA | 16942485 | |
| GO:0045410 | positive regulation of interleukin-6 biosynthetic process | IEA | - | |
| GO:0045429 | positive regulation of nitric oxide biosynthetic process | IDA | 8383325 | |
| GO:0045449 | regulation of transcription | IEA | - | |
| GO:0050796 | regulation of insulin secretion | IDA | 8383325 | |
| GO:0051712 | positive regulation of killing of cells of another organism | IDA | 7544003 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005615 | extracellular space | IEA | - |
Section V. Pathway annotation
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-29 | 475 | 481 | m8 | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.