Gene Page: PDGFB
Summary ?
| GeneID | 5155 |
| Symbol | PDGFB |
| Synonyms | IBGC5|PDGF-2|PDGF2|SIS|SSV|c-sis |
| Description | platelet derived growth factor subunit B |
| Reference | MIM:190040|HGNC:HGNC:8800|Ensembl:ENSG00000100311|HPRD:01815|Vega:OTTHUMG00000151029 |
| Gene type | protein-coding |
| Map location | 22q13.1 |
| Pascal p-value | 0.153 |
| Sherlock p-value | 0.008 |
| Fetal beta | 0.208 |
| eGene | Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
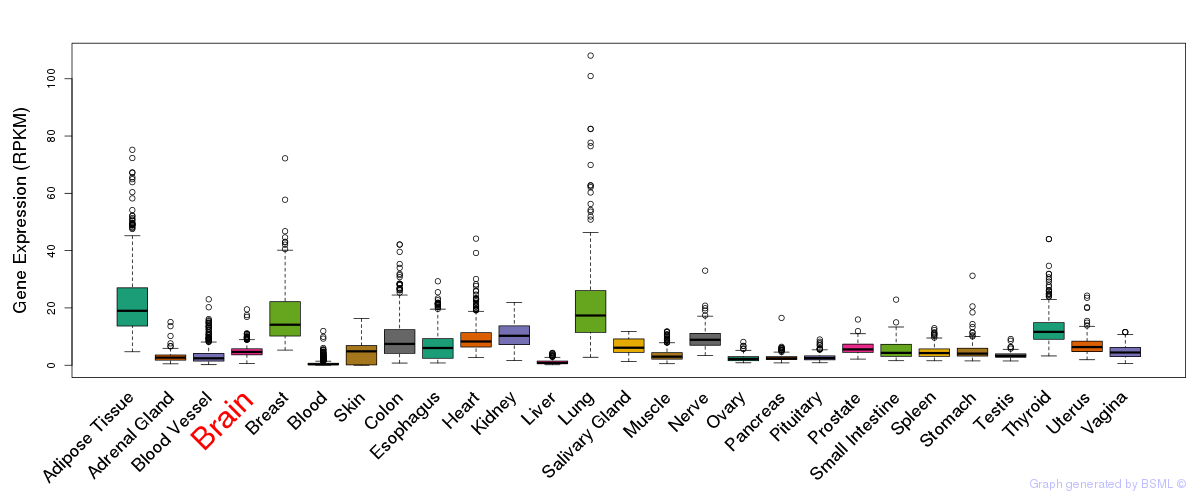
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DUSP7 | 0.77 | 0.74 |
| DUSP8 | 0.76 | 0.75 |
| SPEG | 0.74 | 0.71 |
| DENND1A | 0.73 | 0.75 |
| BAI2 | 0.73 | 0.73 |
| C11orf81 | 0.72 | 0.76 |
| KIAA0664 | 0.72 | 0.73 |
| RHOBTB2 | 0.71 | 0.69 |
| MGAT5B | 0.71 | 0.71 |
| PIP5K1C | 0.71 | 0.70 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SYCP3 | -0.52 | -0.54 |
| RPS27L | -0.51 | -0.52 |
| C1orf54 | -0.49 | -0.49 |
| GNG11 | -0.48 | -0.48 |
| CLEC2B | -0.47 | -0.44 |
| DBI | -0.47 | -0.48 |
| C8orf59 | -0.47 | -0.48 |
| MTCP1NB | -0.46 | -0.48 |
| S100A13 | -0.45 | -0.38 |
| AF347015.21 | -0.45 | -0.43 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005161 | platelet-derived growth factor receptor binding | NAS | 1661670 | |
| GO:0005515 | protein binding | IPI | 17981115 | |
| GO:0008083 | growth factor activity | IEA | - | |
| GO:0048407 | platelet-derived growth factor binding | IPI | 7073684 | |
| GO:0042803 | protein homodimerization activity | IDA | 2836953 | |
| GO:0043498 | cell surface binding | IDA | 291037 |2538439 |2836953 | |
| GO:0046982 | protein heterodimerization activity | IPI | 7073684 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0030031 | cell projection biogenesis | IEA | axon (GO term level: 8) | - |
| GO:0001938 | positive regulation of endothelial cell proliferation | IEA | - | |
| GO:0010544 | negative regulation of platelet activation | IDA | 2538439 | |
| GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process | IDA | 2538439 | |
| GO:0008283 | cell proliferation | IEA | - | |
| GO:0009611 | response to wounding | NAS | 1661670 | |
| GO:0048008 | platelet-derived growth factor receptor signaling pathway | IDA | 2439522 |2536956 |2836953 | |
| GO:0006929 | substrate-bound cell migration | IEA | - | |
| GO:0030036 | actin cytoskeleton organization | IEA | - | |
| GO:0014911 | positive regulation of smooth muscle cell migration | IDA | 9409235 | |
| GO:0050921 | positive regulation of chemotaxis | IDA | 9409235 | |
| GO:0030336 | negative regulation of cell migration | IEA | - | |
| GO:0043406 | positive regulation of MAP kinase activity | IDA | 9685360 | |
| GO:0045740 | positive regulation of DNA replication | IDA | 2439522 |2836953 | |
| GO:0048146 | positive regulation of fibroblast proliferation | IDA | 2439522 | |
| GO:0050730 | regulation of peptidyl-tyrosine phosphorylation | IEA | - | |
| GO:0043536 | positive regulation of blood vessel endothelial cell migration | IDA | 9685360 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | EXP | 287022 | |
| GO:0005576 | extracellular region | NAS | 1661670 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0009986 | cell surface | IDA | 291037 |2836953 |9685360 | |
| GO:0031089 | platelet dense granule lumen | EXP | 287022 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| A2M | CPAMD5 | DKFZp779B086 | FWP007 | S863-7 | alpha-2-macroglobulin | - | HPRD | 10681572 |
| ART1 | ART2 | CD296 | MGC133217 | RT6 | ADP-ribosyltransferase 1 | - | HPRD | 11498506 |
| COL1A1 | OI4 | collagen, type I, alpha 1 | - | HPRD | 10446987 |
| HNRNPC | C1 | C2 | HNRNP | HNRPC | MGC104306 | MGC105117 | MGC117353 | MGC131677 | SNRPC | heterogeneous nuclear ribonucleoprotein C (C1/C2) | - | HPRD | 10409733 |
| HRAS | C-BAS/HAS | C-H-RAS | C-HA-RAS1 | CTLO | H-RASIDX | HAMSV | HRAS1 | K-RAS | N-RAS | RASH1 | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | - | HPRD | 10485711 |
| HSPG2 | PLC | PRCAN | SJA | SJS | SJS1 | heparan sulfate proteoglycan 2 | - | HPRD,BioGRID | 9692901 |
| LRP1 | A2MR | APOER | APR | CD91 | FLJ16451 | IGFBP3R | LRP | MGC88725 | TGFBR5 | low density lipoprotein-related protein 1 (alpha-2-macroglobulin receptor) | - | HPRD,BioGRID | 11854294 |
| PDAP1 | HASPP28 | PAP | PAP1 | PDGFA associated protein 1 | - | HPRD,BioGRID | 8780057 |
| PDGFB | FLJ12858 | PDGF2 | SIS | SSV | c-sis | platelet-derived growth factor beta polypeptide (simian sarcoma viral (v-sis) oncogene homolog) | - | HPRD,BioGRID | 1396586 |
| PDGFRA | CD140A | MGC74795 | PDGFR2 | Rhe-PDGFRA | platelet-derived growth factor receptor, alpha polypeptide | - | HPRD,BioGRID | 2544881 |
| PDGFRB | CD140B | JTK12 | PDGF-R-beta | PDGFR | PDGFR1 | platelet-derived growth factor receptor, beta polypeptide | - | HPRD | 11903042 |
| SPARC | ON | secreted protein, acidic, cysteine-rich (osteonectin) | Reconstituted Complex | BioGRID | 1311092 |
| THBS1 | THBS | TSP | TSP1 | thrombospondin 1 | - | HPRD,BioGRID | 9334164 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION | 267 | 161 | All SZGR 2.0 genes in this pathway |
| KEGG FOCAL ADHESION | 201 | 138 | All SZGR 2.0 genes in this pathway |
| KEGG GAP JUNCTION | 90 | 68 | All SZGR 2.0 genes in this pathway |
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG RENAL CELL CARCINOMA | 70 | 60 | All SZGR 2.0 genes in this pathway |
| KEGG GLIOMA | 65 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG PROSTATE CANCER | 89 | 75 | All SZGR 2.0 genes in this pathway |
| KEGG MELANOMA | 71 | 57 | All SZGR 2.0 genes in this pathway |
| PID PTP1B PATHWAY | 52 | 40 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN3 PATHWAY | 43 | 25 | All SZGR 2.0 genes in this pathway |
| PID S1P S1P3 PATHWAY | 29 | 24 | All SZGR 2.0 genes in this pathway |
| PID NECTIN PATHWAY | 30 | 20 | All SZGR 2.0 genes in this pathway |
| PID TCPTP PATHWAY | 43 | 33 | All SZGR 2.0 genes in this pathway |
| PID SHP2 PATHWAY | 58 | 46 | All SZGR 2.0 genes in this pathway |
| PID S1P S1P1 PATHWAY | 21 | 18 | All SZGR 2.0 genes in this pathway |
| PID PDGFRB PATHWAY | 129 | 103 | All SZGR 2.0 genes in this pathway |
| REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | 89 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY PDGF | 122 | 93 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | 95 | 76 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING DN | 46 | 29 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE UP | 485 | 293 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS GREY DN | 74 | 44 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA UP | 544 | 308 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA UP | 171 | 112 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HIF1A TARGETS DN | 91 | 58 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HIF1A AND HIF2A TARGETS DN | 104 | 72 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN UP | 612 | 367 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS F UP | 185 | 119 | All SZGR 2.0 genes in this pathway |
| STREICHER LSM1 TARGETS UP | 44 | 34 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 3 | 48 | 31 | All SZGR 2.0 genes in this pathway |
| MYLLYKANGAS AMPLIFICATION HOT SPOT 22 | 13 | 9 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| TAGHAVI NEOPLASTIC TRANSFORMATION | 12 | 10 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS DN | 357 | 212 | All SZGR 2.0 genes in this pathway |
| TENEDINI MEGAKARYOCYTE MARKERS | 66 | 48 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA UP | 207 | 145 | All SZGR 2.0 genes in this pathway |
| GALINDO IMMUNE RESPONSE TO ENTEROTOXIN | 85 | 67 | All SZGR 2.0 genes in this pathway |
| KANG IMMORTALIZED BY TERT UP | 89 | 61 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| MOREAUX B LYMPHOCYTE MATURATION BY TACI UP | 92 | 58 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP | 390 | 242 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER HEREDITARY VS SPORADIC | 50 | 32 | All SZGR 2.0 genes in this pathway |
| HARRIS HYPOXIA | 81 | 64 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| LEONARD HYPOXIA | 47 | 35 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER BRCA1 VS BRCA2 | 163 | 113 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN DN | 271 | 175 | All SZGR 2.0 genes in this pathway |
| CLASPER LYMPHATIC VESSELS DURING METASTASIS UP | 20 | 12 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN UP | 439 | 257 | All SZGR 2.0 genes in this pathway |
| SAGIV CD24 TARGETS DN | 46 | 26 | All SZGR 2.0 genes in this pathway |
| LABBE WNT3A TARGETS UP | 112 | 71 | All SZGR 2.0 genes in this pathway |
| LABBE TGFB1 TARGETS UP | 102 | 64 | All SZGR 2.0 genes in this pathway |
| LABBE TARGETS OF TGFB1 AND WNT3A UP | 111 | 70 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA UP | 259 | 185 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA METAGENE | 242 | 168 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS UP | 165 | 118 | All SZGR 2.0 genes in this pathway |
| MIZUKAMI HYPOXIA DN | 6 | 5 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER DN | 238 | 145 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 AND H3K27ME3 | 349 | 234 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN | 253 | 192 | All SZGR 2.0 genes in this pathway |
| DANG MYC TARGETS DN | 31 | 25 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K27ME3 | 341 | 243 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS DN | 315 | 215 | All SZGR 2.0 genes in this pathway |
| KATSANOU ELAVL1 TARGETS UP | 169 | 105 | All SZGR 2.0 genes in this pathway |
| HUANG GATA2 TARGETS DN | 72 | 52 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR UP | 199 | 143 | All SZGR 2.0 genes in this pathway |
| NABA SECRETED FACTORS | 344 | 197 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| let-7/98 | 796 | 802 | m8 | hsa-let-7abrain | UGAGGUAGUAGGUUGUAUAGUU |
| hsa-let-7bbrain | UGAGGUAGUAGGUUGUGUGGUU | ||||
| hsa-let-7cbrain | UGAGGUAGUAGGUUGUAUGGUU | ||||
| hsa-let-7dbrain | AGAGGUAGUAGGUUGCAUAGU | ||||
| hsa-let-7ebrain | UGAGGUAGGAGGUUGUAUAGU | ||||
| hsa-let-7fbrain | UGAGGUAGUAGAUUGUAUAGUU | ||||
| hsa-miR-98brain | UGAGGUAGUAAGUUGUAUUGUU | ||||
| hsa-let-7gSZ | UGAGGUAGUAGUUUGUACAGU | ||||
| hsa-let-7ibrain | UGAGGUAGUAGUUUGUGCUGU | ||||
| miR-29 | 132 | 138 | m8 | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
| miR-496 | 1593 | 1599 | 1A | hsa-miR-496 | AUUACAUGGCCAAUCUC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.