Gene Page: FYN
Summary ?
| GeneID | 2534 |
| Symbol | FYN |
| Synonyms | SLK|SYN|p59-FYN |
| Description | FYN proto-oncogene, Src family tyrosine kinase |
| Reference | MIM:137025|HGNC:HGNC:4037|Ensembl:ENSG00000010810|HPRD:00655|Vega:OTTHUMG00000016305 |
| Gene type | protein-coding |
| Map location | 6q21 |
| Pascal p-value | 0.009 |
| Sherlock p-value | 0.861 |
| Fetal beta | 1.145 |
| DMG | 1 (# studies) |
| eGene | Nucleus accumbens basal ganglia |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Darnell FMRP targets Ascano FMRP targets Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Expression | Meta-analysis of gene expression | P value: 1.398 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 1.777 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg20985014 | 6 | 112374151 | FYN | 0.003 | -5.708 | DMG:vanEijk_2014 |
Section II. Transcriptome annotation
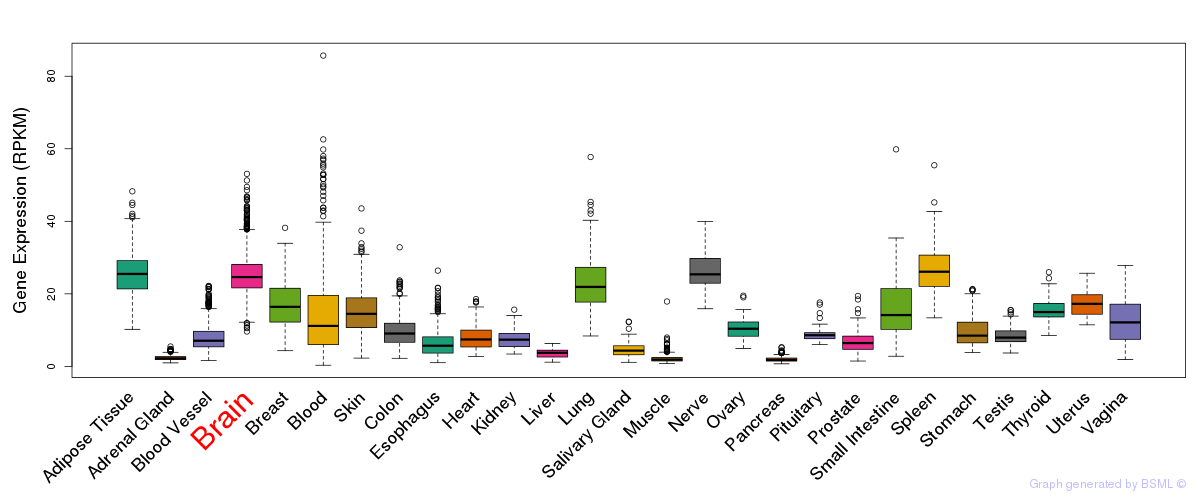
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NDRG4 | 0.86 | 0.89 |
| TTC7B | 0.85 | 0.86 |
| CLPTM1 | 0.85 | 0.83 |
| PLBD2 | 0.84 | 0.81 |
| AP001107.1 | 0.83 | 0.86 |
| CLSTN3 | 0.83 | 0.83 |
| DCTN1 | 0.82 | 0.81 |
| PINK1 | 0.82 | 0.86 |
| C2CD2L | 0.82 | 0.85 |
| ADAMTSL2 | 0.82 | 0.84 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RBMX2 | -0.48 | -0.55 |
| FAM159B | -0.48 | -0.66 |
| BCL7C | -0.46 | -0.57 |
| C21orf57 | -0.44 | -0.53 |
| RPL31 | -0.44 | -0.50 |
| RPL35 | -0.44 | -0.51 |
| RPS20 | -0.43 | -0.55 |
| C9orf46 | -0.43 | -0.50 |
| RPL24 | -0.43 | -0.50 |
| RPL13AP22 | -0.43 | -0.61 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004713 | protein tyrosine kinase activity | IEA | - | |
| GO:0004715 | non-membrane spanning protein tyrosine kinase activity | NAS | 7822789 | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0042802 | identical protein binding | IPI | 14993658 | |
| GO:0030145 | manganese ion binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0006468 | protein amino acid phosphorylation | NAS | 7822789 | |
| GO:0007243 | protein kinase cascade | TAS | 1361685 | |
| GO:0007631 | feeding behavior | TAS | 8264796 | |
| GO:0007612 | learning | TAS | 1361685 | |
| GO:0006816 | calcium ion transport | NAS | 7822789 | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0050852 | T cell receptor signaling pathway | IDA | 7722293 | |
| GO:0044419 | interspecies interaction between organisms | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 10858437 | |
| GO:0005768 | endosome | IDA | 15611048 | |
| GO:0005886 | plasma membrane | IDA | 15611048 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ADAM15 | MDC15 | ADAM metallopeptidase domain 15 | - | HPRD,BioGRID | 11741929 |
| ADD2 | ADDB | adducin 2 (beta) | - | HPRD,BioGRID | 11526103 |
| BCAR1 | CAS | CAS1 | CASS1 | CRKAS | P130Cas | breast cancer anti-estrogen resistance 1 | - | HPRD,BioGRID | 9188452 |
| BCL3 | BCL4 | D19S37 | B-cell CLL/lymphoma 3 | - | HPRD,BioGRID | 9576921 |
| BTK | AGMX1 | AT | ATK | BPK | IMD1 | MGC126261 | MGC126262 | PSCTK1 | XLA | Bruton agammaglobulinemia tyrosine kinase | Reconstituted Complex | BioGRID | 8058772 |
| CBL | C-CBL | CBL2 | RNF55 | Cas-Br-M (murine) ecotropic retroviral transforming sequence | - | HPRD,BioGRID | 9535867 |
| CBLC | CBL-3 | CBL-SL | RNF57 | Cas-Br-M (murine) ecotropic retroviral transforming sequence c | Affinity Capture-Western Reconstituted Complex | BioGRID | 10571044 |
| CD19 | B4 | MGC12802 | CD19 molecule | - | HPRD | 7589101 |
| CD2 | SRBC | T11 | CD2 molecule | - | HPRD,BioGRID | 9677430 |
| CD226 | DNAM-1 | DNAM1 | PTA1 | TLiSA1 | CD226 molecule | - | HPRD,BioGRID | 10591186 |
| CD2AP | CMS | DKFZp586H0519 | CD2-associated protein | - | HPRD | 10339567 |
| CD36 | CHDS7 | FAT | GP3B | GP4 | GPIV | PASIV | SCARB3 | CD36 molecule (thrombospondin receptor) | - | HPRD,BioGRID | 7521304 |
| CD44 | CDW44 | CSPG8 | ECMR-III | HCELL | IN | LHR | MC56 | MDU2 | MDU3 | MGC10468 | MIC4 | MUTCH-I | Pgp1 | CD44 molecule (Indian blood group) | - | HPRD,BioGRID | 9573028 |
| CD48 | BCM1 | BLAST | BLAST1 | MEM-102 | SLAMF2 | hCD48 | mCD48 | CD48 molecule | - | HPRD | 8104794 |
| CD55 | CR | CROM | DAF | TC | CD55 molecule, decay accelerating factor for complement (Cromer blood group) | - | HPRD | 1385527 |
| CDC2 | CDC28A | CDK1 | DKFZp686L20222 | MGC111195 | cell division cycle 2, G1 to S and G2 to M | - | HPRD,BioGRID | 8910336 |
| CDH1 | Arc-1 | CD324 | CDHE | ECAD | LCAM | UVO | cadherin 1, type 1, E-cadherin (epithelial) | Affinity Capture-Western Reconstituted Complex | BioGRID | 12640114 |
| CHRNA7 | CHRNA7-2 | NACHRA7 | cholinergic receptor, nicotinic, alpha 7 | Affinity Capture-Western | BioGRID | 11278378 |
| CNTN1 | F3 | GP135 | contactin 1 | - | HPRD,BioGRID | 7595520 |
| CSF1R | C-FMS | CD115 | CSFR | FIM2 | FMS | colony stimulating factor 1 receptor | - | HPRD,BioGRID | 7681396 |
| CSK | MGC117393 | c-src tyrosine kinase | - | HPRD,BioGRID | 8262983 |
| CTLA4 | CD152 | CELIAC3 | CTLA-4 | GSE | IDDM12 | cytotoxic T-lymphocyte-associated protein 4 | - | HPRD,BioGRID | 9712716 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | Biochemical Activity | BioGRID | 12640114 |
| CTNND1 | CAS | CTNND | KIAA0384 | P120CAS | P120CTN | p120 | catenin (cadherin-associated protein), delta 1 | Affinity Capture-Western Biochemical Activity | BioGRID | 12640114 |12835311 |
| CTNND2 | GT24 | NPRAP | catenin (cadherin-associated protein), delta 2 (neural plakophilin-related arm-repeat protein) | - | HPRD | 12835311 |
| DAG1 | 156DAG | A3a | AGRNR | DAG | dystroglycan 1 (dystrophin-associated glycoprotein 1) | Affinity Capture-Western Reconstituted Complex | BioGRID | 11724572 |
| DCC | CRC18 | CRCR1 | deleted in colorectal carcinoma | DCC interacts with Fyn. | BIND | 15494732 |
| DLG4 | FLJ97752 | FLJ98574 | PSD95 | SAP90 | discs, large homolog 4 (Drosophila) | - | HPRD,BioGRID | 12419528 |
| DOK3 | DOKL | FLJ22570 | FLJ39939 | docking protein 3 | - | HPRD | 10733577 |
| DOK4 | FLJ10488 | docking protein 4 | IRS5/DOK4 interacts with Fyn. This interaction was modelled on a demonstrated interaction between human IRS5/DOK4 and hamster Fyn. | BIND | 12730241 |
| DOK4 | FLJ10488 | docking protein 4 | - | HPRD | 12730241 |
| EPHA4 | HEK8 | SEK | TYRO1 | EPH receptor A4 | - | HPRD,BioGRID | 12084815 |
| EPHA8 | EEK | HEK3 | KIAA1459 | EPH receptor A8 | - | HPRD,BioGRID | 10498895 |
| EPHB3 | ETK2 | HEK2 | TYRO6 | EPH receptor B3 | - | HPRD,BioGRID | 9674711 |
| EVL | RNB6 | Enah/Vasp-like | - | HPRD | 10945997 |
| FAS | ALPS1A | APO-1 | APT1 | CD95 | FAS1 | FASTM | TNFRSF6 | Fas (TNF receptor superfamily, member 6) | - | HPRD,BioGRID | 8626376 |
| FASLG | APT1LG1 | CD178 | CD95L | FASL | TNFSF6 | Fas ligand (TNF superfamily, member 6) | - | HPRD,BioGRID | 7589480 |
| FNBP4 | DKFZp686M14102 | DKFZp779I1064 | FBP30 | FLJ41904 | KIAA1014 | formin binding protein 4 | - | HPRD,BioGRID | 10744724 |
| FYB | ADAP | PRO0823 | SLAP-130 | FYN binding protein (FYB-120/130) | - | HPRD | 7504057 |9207119 |
| FYB | ADAP | PRO0823 | SLAP-130 | FYN binding protein (FYB-120/130) | - | HPRD,BioGRID | 7504057 |
| FYN | MGC45350 | SLK | SYN | FYN oncogene related to SRC, FGR, YES | - | HPRD,BioGRID | 7687536 |
| GAB3 | - | GRB2-associated binding protein 3 | - | HPRD,BioGRID | 11739737 |
| GNB2L1 | Gnb2-rs1 | H12.3 | HLC-7 | PIG21 | RACK1 | guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 | - | HPRD,BioGRID | 11943848 |12524444 |
| GP6 | GPIV | GPVI | MGC138168 | glycoprotein VI (platelet) | - | HPRD,BioGRID | 11943772 |
| GRB10 | GRB-IR | Grb-10 | IRBP | KIAA0207 | MEG1 | RSS | growth factor receptor-bound protein 10 | - | HPRD,BioGRID | 10871840 |
| GRIN2A | NMDAR2A | NR2A | glutamate receptor, ionotropic, N-methyl D-aspartate 2A | Affinity Capture-Western Reconstituted Complex | BioGRID | 9892651 |10458595 |12419528 |12932824 |
| GRIN2A | NMDAR2A | NR2A | glutamate receptor, ionotropic, N-methyl D-aspartate 2A | - | HPRD | 10458595 |12932824 |
| GRIN2B | MGC142178 | MGC142180 | NMDAR2B | NR2B | hNR3 | glutamate receptor, ionotropic, N-methyl D-aspartate 2B | - | HPRD,BioGRID | 10458595 |
| HNRNPK | CSBP | FLJ41122 | HNRPK | TUNP | heterogeneous nuclear ribonucleoprotein K | Fyn interacts with hnRNPK. This interaction was modeled on a demonstrated interaction between Fyn from an unspecified source and human hnRNPK. | BIND | 8810341 |
| IL7R | CD127 | CDW127 | IL-7R-alpha | IL7RA | ILRA | interleukin 7 receptor | - | HPRD,BioGRID | 7515933 |
| ITK | EMT | LYK | MGC126257 | MGC126258 | PSCTK2 | IL2-inducible T-cell kinase | - | HPRD,BioGRID | 8810341 |
| JAK2 | JTK10 | Janus kinase 2 (a protein tyrosine kinase) | - | HPRD,BioGRID | 10551884 |
| KHDRBS1 | FLJ34027 | Sam68 | p62 | KH domain containing, RNA binding, signal transduction associated 1 | - | HPRD,BioGRID | 9045636 |
| MAG | GMA | S-MAG | SIGLEC-4A | SIGLEC4A | myelin associated glycoprotein | - | HPRD,BioGRID | 7525550 |
| MAP2 | DKFZp686I2148 | MAP2A | MAP2B | MAP2C | microtubule-associated protein 2 | Affinity Capture-Western | BioGRID | 11546790 |
| MAPT | DDPAC | FLJ31424 | FTDP-17 | MAPTL | MGC138549 | MSTD | MTBT1 | MTBT2 | PPND | TAU | microtubule-associated protein tau | - | HPRD,BioGRID | 11826099 |
| MCAM | CD146 | MUC18 | melanoma cell adhesion molecule | - | HPRD,BioGRID | 9756930 |
| MS4A1 | B1 | Bp35 | CD20 | LEU-16 | MGC3969 | MS4A2 | S7 | membrane-spanning 4-domains, subfamily A, member 1 | - | HPRD,BioGRID | 7545683 |
| NCAM1 | CD56 | MSK39 | NCAM | neural cell adhesion molecule 1 | - | HPRD | 9079653 |11839780 |
| NEDD9 | CAS-L | CAS2 | CASL | CASS2 | HEF1 | dJ49G10.2 | dJ761I2.1 | neural precursor cell expressed, developmentally down-regulated 9 | - | HPRD,BioGRID | 9360983 |
| NMT1 | NMT | N-myristoyltransferase 1 | Biochemical Activity | BioGRID | 11594778 |
| NPHS1 | CNF | NPHN | nephrosis 1, congenital, Finnish type (nephrin) | - | HPRD,BioGRID | 12668668 |12846735 |
| NTRK2 | GP145-TrkB | TRKB | neurotrophic tyrosine kinase, receptor, type 2 | - | HPRD,BioGRID | 9648856 |
| PAG1 | CBP | FLJ37858 | MGC138364 | PAG | phosphoprotein associated with glycosphingolipid microdomains 1 | - | HPRD,BioGRID | 10790433 |
| PDE4D | DPDE3 | HSPDE4D | PDE4DN2 | STRK1 | phosphodiesterase 4D, cAMP-specific (phosphodiesterase E3 dunce homolog, Drosophila) | PDE4D4 interacts with Fyn SH3 domain. | BIND | 10571082 |
| PDGFRB | CD140B | JTK12 | PDGF-R-beta | PDGFR | PDGFR1 | platelet-derived growth factor receptor, beta polypeptide | - | HPRD | 1696179 |
| PECAM1 | CD31 | PECAM-1 | platelet/endothelial cell adhesion molecule | - | HPRD,BioGRID | 10858437 |
| PIK3R1 | GRB1 | p85 | p85-ALPHA | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | - | HPRD | 8394019 |
| PIK3R1 | GRB1 | p85 | p85-ALPHA | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | Reconstituted Complex | BioGRID | 8294442 |
| PIK3R2 | P85B | p85 | p85-BETA | phosphoinositide-3-kinase, regulatory subunit 2 (beta) | - | HPRD,BioGRID | 1334406 |
| PLAUR | CD87 | UPAR | URKR | plasminogen activator, urokinase receptor | - | HPRD | 9417082 |
| PLCG1 | PLC-II | PLC1 | PLC148 | PLCgamma1 | phospholipase C, gamma 1 | - | HPRD | 10586033 |
| PLCG2 | - | phospholipase C, gamma 2 (phosphatidylinositol-specific) | - | HPRD,BioGRID | 8395016 |
| PLD2 | - | phospholipase D2 | - | HPRD | 12697812 |
| PRKCQ | MGC126514 | MGC141919 | PRKCT | nPKC-theta | protein kinase C, theta | - | HPRD,BioGRID | 10383400 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | - | HPRD,BioGRID | 11278857 |
| PTK2B | CADTK | CAKB | FADK2 | FAK2 | FRNK | PKB | PTK | PYK2 | RAFTK | PTK2B protein tyrosine kinase 2 beta | - | HPRD,BioGRID | 9091579 |9104812 |10867021 |
| PTPN11 | BPTP3 | CFC | MGC14433 | NS1 | PTP-1D | PTP2C | SH-PTP2 | SH-PTP3 | SHP2 | protein tyrosine phosphatase, non-receptor type 11 | - | HPRD,BioGRID | 10212213 |
| PTPN5 | FLJ14427 | PTPSTEP | STEP | protein tyrosine phosphatase, non-receptor type 5 (striatum-enriched) | - | HPRD,BioGRID | 11983687 |
| PTPRA | HEPTP | HLPR | HPTPA | HPTPalpha | LRP | PTPA | PTPRL2 | R-PTP-alpha | RPTPA | protein tyrosine phosphatase, receptor type, A | - | HPRD,BioGRID | 9535845 |
| PTPRC | B220 | CD45 | CD45R | GP180 | LCA | LY5 | T200 | protein tyrosine phosphatase, receptor type, C | - | HPRD | 9368621 |12589038 |
| PXN | FLJ16691 | paxillin | - | HPRD,BioGRID | 10804218 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | - | HPRD,BioGRID | 7517401 |
| RICS | GC-GAP | GRIT | KIAA0712 | MGC1892 | p200RhoGAP | p250GAP | Rho GTPase-activating protein | - | HPRD,BioGRID | 12788081 |12857875 |
| RPL10 | DKFZp686J1851 | DXS648 | DXS648E | FLJ23544 | FLJ27072 | NOV | QM | ribosomal protein L10 | Reconstituted Complex | BioGRID | 12138090 |
| SDC3 | N-syndecan | SDCN | SYND3 | syndecan 3 | - | HPRD,BioGRID | 9553134 |
| SH2B2 | APS | SH2B adaptor protein 2 | - | HPRD | 10872802 |
| SH2D1A | DSHP | EBVS | FLJ18687 | FLJ92177 | IMD5 | LYP | MTCP1 | SAP | XLP | XLPD | SH2 domain protein 1A | - | HPRD,BioGRID | 12545174 |
| SH3BP2 | 3BP2 | CRBM | CRPM | FLJ42079 | RES4-23 | SH3-domain binding protein 2 | Two-hybrid | BioGRID | 9846481 |
| SIT1 | MGC125908 | MGC125909 | MGC125910 | RP11-331F9.5 | SIT | signaling threshold regulating transmembrane adaptor 1 | - | HPRD,BioGRID | 10209036 |
| SKAP1 | SCAP1 | SKAP55 | src kinase associated phosphoprotein 1 | - | HPRD,BioGRID | 9195899 |
| SKAP2 | MGC10411 | MGC33304 | PRAP | RA70 | SAPS | SCAP2 | SKAP-HOM | SKAP55R | src kinase associated phosphoprotein 2 | - | HPRD,BioGRID | 9837776 |
| SLAMF1 | CD150 | CDw150 | SLAM | signaling lymphocytic activation molecule family member 1 | Reconstituted Complex | BioGRID | 12545174 |
| SNCA | MGC110988 | NACP | PARK1 | PARK4 | PD1 | synuclein, alpha (non A4 component of amyloid precursor) | - | HPRD,BioGRID | 11162638 |
| SOCS1 | CIS1 | CISH1 | JAB | SOCS-1 | SSI-1 | SSI1 | TIP3 | suppressor of cytokine signaling 1 | Socs1 interacts with Fyn. This interaction was modeled on a demonstrated interaction between mouse Socs1 and Fyn from an unspecified species. | BIND | 10022833 |
| SOCS1 | CIS1 | CISH1 | JAB | SOCS-1 | SSI-1 | SSI1 | TIP3 | suppressor of cytokine signaling 1 | - | HPRD,BioGRID | 10022833 |
| SOS1 | GF1 | GGF1 | GINGF | HGF | NS4 | son of sevenless homolog 1 (Drosophila) | - | HPRD,BioGRID | 9856337 |
| SPN | CD43 | GPL115 | LSN | sialophorin | - | HPRD | 9603925 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | Protein-peptide | BioGRID | 9169421 |
| SYK | DKFZp313N1010 | FLJ25043 | FLJ37489 | spleen tyrosine kinase | - | HPRD,BioGRID | 9535867 |
| THY1 | CD90 | FLJ33325 | Thy-1 cell surface antigen | - | HPRD,BioGRID | 1351058 |
| TNK2 | ACK | ACK1 | FLJ44758 | FLJ45547 | p21cdc42Hs | tyrosine kinase, non-receptor, 2 | - | HPRD,BioGRID | 11087735 |
| TOM1L1 | OK/KNS-CL.3 | SRCASM | target of myb1 (chicken)-like 1 | Reconstituted Complex | BioGRID | 11711534 |
| TRAT1 | HSPC062 | TCRIM | TRIM | T cell receptor associated transmembrane adaptor 1 | Reconstituted Complex | BioGRID | 10790433 |
| TRPC6 | FLJ11098 | FLJ14863 | FSGS2 | TRP6 | transient receptor potential cation channel, subfamily C, member 6 | - | HPRD,BioGRID | 14761972 |
| TRPV4 | OTRPC4 | TRP12 | VR-OAC | VRL-2 | VRL2 | VROAC | transient receptor potential cation channel, subfamily V, member 4 | - | HPRD,BioGRID | 12538589 |
| TUBA1B | K-ALPHA-1 | tubulin, alpha 1b | - | HPRD | 11826099 |
| TUBA3C | TUBA2 | bA408E5.3 | tubulin, alpha 3c | - | HPRD,BioGRID | 8566014 |11826099 |
| TUBA4A | FLJ30169 | H2-ALPHA | TUBA1 | tubulin, alpha 4a | - | HPRD | 8566014 |
| TYK2 | JTK1 | tyrosine kinase 2 | - | HPRD,BioGRID | 9196040 |
| TYRO3 | BYK | Brt | Dtk | FLJ16467 | RSE | Sky | Tif | TYRO3 protein tyrosine kinase | - | HPRD | 7537495 |
| UNC119 | HRG4 | unc-119 homolog (C. elegans) | - | HPRD,BioGRID | 14757743 |
| VAV1 | VAV | vav 1 guanine nucleotide exchange factor | - | HPRD,BioGRID | 9822663 |
| WAS | IMD2 | THC | WASP | Wiskott-Aldrich syndrome (eczema-thrombocytopenia) | Affinity Capture-Western Reconstituted Complex | BioGRID | 8805332 |10532312 |
| WAS | IMD2 | THC | WASP | Wiskott-Aldrich syndrome (eczema-thrombocytopenia) | - | HPRD | 8805332 |10224664 |10532312 |
| WASF1 | FLJ31482 | KIAA0269 | SCAR1 | WAVE | WAVE1 | WAS protein family, member 1 | - | HPRD | 10532312 |
| WASF2 | SCAR2 | WAVE2 | dJ393P12.2 | WAS protein family, member 2 | - | HPRD | 10532312 |
| YES1 | HsT441 | P61-YES | Yes | c-yes | v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 | Affinity Capture-Western | BioGRID | 12640114 |
| YTHDC1 | KIAA1966 | YT521 | YT521-B | YTH domain containing 1 | - | HPRD,BioGRID | 15175272 |
| ZAP70 | FLJ17670 | FLJ17679 | SRK | STD | TZK | ZAP-70 | zeta-chain (TCR) associated protein kinase 70kDa | - | HPRD,BioGRID | 7760813 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG AXON GUIDANCE | 129 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG FOCAL ADHESION | 201 | 138 | All SZGR 2.0 genes in this pathway |
| KEGG ADHERENS JUNCTION | 75 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG NATURAL KILLER CELL MEDIATED CYTOTOXICITY | 137 | 92 | All SZGR 2.0 genes in this pathway |
| KEGG T CELL RECEPTOR SIGNALING PATHWAY | 108 | 89 | All SZGR 2.0 genes in this pathway |
| KEGG FC EPSILON RI SIGNALING PATHWAY | 79 | 58 | All SZGR 2.0 genes in this pathway |
| KEGG PRION DISEASES | 35 | 28 | All SZGR 2.0 genes in this pathway |
| KEGG PATHOGENIC ESCHERICHIA COLI INFECTION | 59 | 36 | All SZGR 2.0 genes in this pathway |
| KEGG VIRAL MYOCARDITIS | 73 | 58 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BIOPEPTIDES PATHWAY | 45 | 35 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EPHA4 PATHWAY | 10 | 8 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ECM PATHWAY | 24 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL7 PATHWAY | 17 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA INTEGRIN PATHWAY | 38 | 29 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TCRA PATHWAY | 13 | 10 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TCR PATHWAY | 49 | 37 | All SZGR 2.0 genes in this pathway |
| ST INTEGRIN SIGNALING PATHWAY | 82 | 62 | All SZGR 2.0 genes in this pathway |
| PID FCER1 PATHWAY | 62 | 43 | All SZGR 2.0 genes in this pathway |
| PID ERBB4 PATHWAY | 38 | 32 | All SZGR 2.0 genes in this pathway |
| PID GLYPICAN 1PATHWAY | 27 | 20 | All SZGR 2.0 genes in this pathway |
| PID TCR PATHWAY | 66 | 51 | All SZGR 2.0 genes in this pathway |
| PID PTP1B PATHWAY | 52 | 40 | All SZGR 2.0 genes in this pathway |
| PID REELIN PATHWAY | 29 | 29 | All SZGR 2.0 genes in this pathway |
| PID P38 ALPHA BETA PATHWAY | 31 | 25 | All SZGR 2.0 genes in this pathway |
| PID CD8 TCR PATHWAY | 53 | 42 | All SZGR 2.0 genes in this pathway |
| PID ANGIOPOIETIN RECEPTOR PATHWAY | 50 | 41 | All SZGR 2.0 genes in this pathway |
| PID TXA2PATHWAY | 57 | 43 | All SZGR 2.0 genes in this pathway |
| PID NETRIN PATHWAY | 32 | 27 | All SZGR 2.0 genes in this pathway |
| PID IL2 1PATHWAY | 55 | 43 | All SZGR 2.0 genes in this pathway |
| PID CXCR4 PATHWAY | 102 | 78 | All SZGR 2.0 genes in this pathway |
| PID PI3KCI PATHWAY | 49 | 40 | All SZGR 2.0 genes in this pathway |
| PID AJDISS 2PATHWAY | 48 | 38 | All SZGR 2.0 genes in this pathway |
| PID AMB2 NEUTROPHILS PATHWAY | 41 | 32 | All SZGR 2.0 genes in this pathway |
| PID EPHA FWDPATHWAY | 34 | 29 | All SZGR 2.0 genes in this pathway |
| PID ECADHERIN KERATINOCYTE PATHWAY | 21 | 19 | All SZGR 2.0 genes in this pathway |
| PID PDGFRB PATHWAY | 129 | 103 | All SZGR 2.0 genes in this pathway |
| PID NEPHRIN NEPH1 PATHWAY | 31 | 24 | All SZGR 2.0 genes in this pathway |
| PID VEGFR1 2 PATHWAY | 69 | 57 | All SZGR 2.0 genes in this pathway |
| PID SYNDECAN 3 PATHWAY | 17 | 15 | All SZGR 2.0 genes in this pathway |
| PID EPHRINB REV PATHWAY | 30 | 25 | All SZGR 2.0 genes in this pathway |
| PID ALPHA SYNUCLEIN PATHWAY | 33 | 25 | All SZGR 2.0 genes in this pathway |
| PID FAK PATHWAY | 59 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY SCF KIT | 78 | 59 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CELL COMMUNICATION | 120 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF KIT SIGNALING | 17 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB2 | 101 | 78 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | 29 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | 126 | 90 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | 91 | 65 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | 12 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME PECAM1 INTERACTIONS | 10 | 7 | All SZGR 2.0 genes in this pathway |
| REACTOME GPVI MEDIATED ACTIVATION CASCADE | 31 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME AXON GUIDANCE | 251 | 188 | All SZGR 2.0 genes in this pathway |
| REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | 64 | 49 | All SZGR 2.0 genes in this pathway |
| REACTOME CD28 CO STIMULATION | 32 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME CRMPS IN SEMA3A SIGNALING | 14 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | 15 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME CD28 DEPENDENT VAV1 PATHWAY | 11 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME SEMAPHORIN INTERACTIONS | 68 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | 13 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME COSTIMULATION BY THE CD28 FAMILY | 63 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | 22 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME CTLA4 INHIBITORY SIGNALING | 21 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ILS | 107 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME NETRIN1 SIGNALING | 41 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | 13 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF SIGNALING BY CBL | 18 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME NEPHRIN INTERACTIONS | 20 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME IL 3 5 AND GM CSF SIGNALING | 43 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV INFECTION | 207 | 122 | All SZGR 2.0 genes in this pathway |
| REACTOME HOST INTERACTIONS OF HIV FACTORS | 132 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | 28 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| LIU PROSTATE CANCER DN | 481 | 290 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE UP | 351 | 230 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA DN | 320 | 184 | All SZGR 2.0 genes in this pathway |
| LIU VMYB TARGETS UP | 127 | 78 | All SZGR 2.0 genes in this pathway |
| RODRIGUES DCC TARGETS DN | 121 | 84 | All SZGR 2.0 genes in this pathway |
| BILBAN B CLL LPL DN | 42 | 25 | All SZGR 2.0 genes in this pathway |
| AKL HTLV1 INFECTION DN | 68 | 41 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA UP | 171 | 112 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA BY DMOG UP | 130 | 85 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HIF1A TARGETS DN | 91 | 58 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HIF1A AND HIF2A TARGETS DN | 104 | 72 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS UP | 149 | 84 | All SZGR 2.0 genes in this pathway |
| PROVENZANI METASTASIS UP | 194 | 112 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 DN | 378 | 231 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2B | 392 | 251 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER HIGH RECURRENCE | 49 | 31 | All SZGR 2.0 genes in this pathway |
| LUI THYROID CANCER CLUSTER 1 | 51 | 33 | All SZGR 2.0 genes in this pathway |
| XU HGF SIGNALING NOT VIA AKT1 48HR UP | 35 | 20 | All SZGR 2.0 genes in this pathway |
| WANG METHYLATED IN BREAST CANCER | 35 | 25 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS NEUROEPITHELIUM DN | 164 | 111 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| WU CELL MIGRATION | 184 | 114 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| DING LUNG CANCER MUTATED FREQUENTLY | 12 | 10 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA CD1 UP | 45 | 29 | All SZGR 2.0 genes in this pathway |
| NADLER HYPERGLYCEMIA AT OBESITY | 58 | 35 | All SZGR 2.0 genes in this pathway |
| KUMAR TARGETS OF MLL AF9 FUSION | 405 | 264 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP | 390 | 242 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN UP | 262 | 186 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR DN | 178 | 121 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK DN | 318 | 220 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN WITHOUT MGMT 48HR DN | 32 | 25 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| YIH RESPONSE TO ARSENITE C3 | 36 | 24 | All SZGR 2.0 genes in this pathway |
| DAZARD UV RESPONSE CLUSTER G6 | 153 | 112 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 48HR DN | 161 | 105 | All SZGR 2.0 genes in this pathway |
| SAFFORD T LYMPHOCYTE ANERGY | 87 | 54 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D8 | 40 | 29 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN DN | 271 | 175 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 12HR DN | 101 | 64 | All SZGR 2.0 genes in this pathway |
| KRIGE AMINO ACID DEPRIVATION | 29 | 20 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| WALLACE PROSTATE CANCER RACE UP | 299 | 167 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE DN | 315 | 197 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL OPTIMAL DEBULKING | 246 | 152 | All SZGR 2.0 genes in this pathway |
| PODAR RESPONSE TO ADAPHOSTIN UP | 147 | 98 | All SZGR 2.0 genes in this pathway |
| LU TUMOR ANGIOGENESIS UP | 25 | 22 | All SZGR 2.0 genes in this pathway |
| FIRESTEIN PROLIFERATION | 175 | 125 | All SZGR 2.0 genes in this pathway |
| JIANG TIP30 TARGETS UP | 46 | 28 | All SZGR 2.0 genes in this pathway |
| GOLDRATH ANTIGEN RESPONSE | 346 | 192 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 DN | 315 | 201 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF DN | 235 | 144 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 11 | 36 | 24 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE DN | 181 | 97 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G123 DN | 51 | 30 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| NAKAMURA ADIPOGENESIS EARLY UP | 66 | 44 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G2 M | 216 | 124 | All SZGR 2.0 genes in this pathway |
| HIRSCH CELLULAR TRANSFORMATION SIGNATURE UP | 242 | 159 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS UP | 259 | 159 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION DN | 321 | 200 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN | 448 | 282 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| ALFANO MYC TARGETS | 239 | 156 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-27 | 186 | 192 | 1A | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-369-3p | 387 | 393 | m8 | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-96 | 295 | 301 | m8 | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.