Gene Page: CCND1
Summary ?
| GeneID | 595 |
| Symbol | CCND1 |
| Synonyms | BCL1|D11S287E|PRAD1|U21B31 |
| Description | cyclin D1 |
| Reference | MIM:168461|HGNC:HGNC:1582|Ensembl:ENSG00000110092|HPRD:01346|Vega:OTTHUMG00000167877 |
| Gene type | protein-coding |
| Map location | 11q13 |
| Pascal p-value | 0.591 |
| Sherlock p-value | 0.045 |
| Fetal beta | -0.397 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.015 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg05200606 | 11 | 69457819 | CCND1 | 1.42E-8 | -0.017 | 5.5E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs11708509 | chr3 | 98075542 | CCND1 | 595 | 0.18 | trans |
Section II. Transcriptome annotation
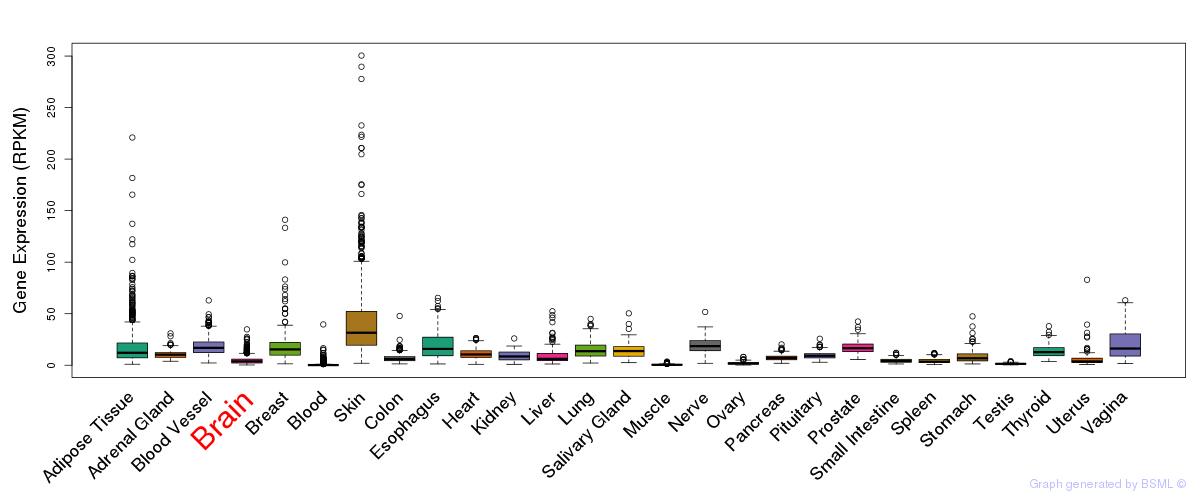
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ARFGAP2 | 0.89 | 0.87 |
| VPS26B | 0.89 | 0.89 |
| VPS4A | 0.88 | 0.85 |
| RNF187 | 0.88 | 0.84 |
| GDI1 | 0.88 | 0.83 |
| L3MBTL2 | 0.86 | 0.83 |
| PPP2R2D | 0.85 | 0.82 |
| TBC1D10B | 0.85 | 0.83 |
| CSRP2BP | 0.85 | 0.80 |
| METTL13 | 0.85 | 0.83 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.76 | -0.74 |
| AF347015.31 | -0.75 | -0.74 |
| AF347015.8 | -0.72 | -0.73 |
| AF347015.21 | -0.72 | -0.70 |
| AF347015.33 | -0.72 | -0.69 |
| AF347015.2 | -0.71 | -0.69 |
| HIGD1B | -0.71 | -0.73 |
| MT-CYB | -0.71 | -0.70 |
| FXYD1 | -0.71 | -0.73 |
| AF347015.26 | -0.70 | -0.68 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 15232106 |17254966 |17274640 |17334399 | |
| GO:0004672 | protein kinase activity | IEA | - | |
| GO:0016538 | cyclin-dependent protein kinase regulator activity | IEA | - | |
| GO:0019901 | protein kinase binding | IPI | 8114739 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000082 | G1/S transition of mitotic cell cycle | NAS | - | |
| GO:0000320 | re-entry into mitotic cell cycle | IEA | - | |
| GO:0001934 | positive regulation of protein amino acid phosphorylation | IDA | 8114739 | |
| GO:0030968 | endoplasmic reticulum unfolded protein response | IEA | - | |
| GO:0051301 | cell division | IEA | - | |
| GO:0045737 | positive regulation of cyclin-dependent protein kinase activity | IDA | 8114739 | |
| GO:0045444 | fat cell differentiation | IEA | - | |
| GO:0051726 | regulation of cell cycle | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000307 | cyclin-dependent protein kinase holoenzyme complex | IEA | - | |
| GO:0005829 | cytosol | EXP | 9106657 | |
| GO:0005829 | cytosol | IEA | - | |
| GO:0005575 | cellular_component | ND | - | |
| GO:0005622 | intracellular | IDA | 11256614 | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005654 | nucleoplasm | EXP | 9190208 |16782892 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AKAP8 | AKAP95 | DKFZp586B1222 | A kinase (PRKA) anchor protein 8 | - | HPRD,BioGRID | 14641107 |
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | - | HPRD,BioGRID | 10344732 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | - | HPRD,BioGRID | 9244350 |
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | BRCA1 associates with Cyclin D1 | BIND | 9244350 |
| CALM1 | CALML2 | CAMI | DD132 | PHKD | calmodulin 1 (phosphorylase kinase, delta) | - | HPRD | 9837900 |
| CCNDBP1 | DIP1 | GCIP | cyclin D-type binding-protein 1 | - | HPRD,BioGRID | 10801854 |
| CDK4 | CMM3 | MGC14458 | PSK-J3 | cyclin-dependent kinase 4 | - | HPRD | 9150368 |9837900 |
| CDK4 | CMM3 | MGC14458 | PSK-J3 | cyclin-dependent kinase 4 | CDK4 interacts with Cyclin D1. This interaction was modelled on a demonstrated interaction between human CDK4 and Cyclin D1 from an unspecified species. | BIND | 9228064 |
| CDK4 | CMM3 | MGC14458 | PSK-J3 | cyclin-dependent kinase 4 | CCND1 (cyclin D1) interacts with CDK4. | BIND | 8662825 |
| CDK4 | CMM3 | MGC14458 | PSK-J3 | cyclin-dependent kinase 4 | Affinity Capture-Western Reconstituted Complex | BioGRID | 8259215 |9837900 |10580009 |11360184 |
| CDK6 | MGC59692 | PLSTIRE | STQTL11 | cyclin-dependent kinase 6 | Affinity Capture-Western Reconstituted Complex | BioGRID | 10580009 |11360184 |
| CDK6 | MGC59692 | PLSTIRE | STQTL11 | cyclin-dependent kinase 6 | - | HPRD | 11739795 |
| CDK6 | MGC59692 | PLSTIRE | STQTL11 | cyclin-dependent kinase 6 | CCND1 (cyclin D1) interacts with CDK6. | BIND | 12867429 |
| CDKN1A | CAP20 | CDKN1 | CIP1 | MDA-6 | P21 | SDI1 | WAF1 | p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | - | HPRD,BioGRID | 7478582 |8657154 |
| CDKN1A | CAP20 | CDKN1 | CIP1 | MDA-6 | P21 | SDI1 | WAF1 | p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | p21 interacts with cyclin D. | BIND | 9632134 |
| CDKN1A | CAP20 | CDKN1 | CIP1 | MDA-6 | P21 | SDI1 | WAF1 | p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | Cyclin D1 interacts with p21. | BIND | 8999999 |
| CDKN1A | CAP20 | CDKN1 | CIP1 | MDA-6 | P21 | SDI1 | WAF1 | p21CIP1 | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | CDKN1A (p21) interacts with CCND1 (cyclin D1). | BIND | 8662825 |
| CDKN1B | CDKN4 | KIP1 | MEN1B | MEN4 | P27KIP1 | cyclin-dependent kinase inhibitor 1B (p27, Kip1) | in vitro Reconstituted Complex | BioGRID | 10908655 |11360184 |
| CDKN1B | CDKN4 | KIP1 | MEN1B | MEN4 | P27KIP1 | cyclin-dependent kinase inhibitor 1B (p27, Kip1) | p27 interacts with cyclin D. | BIND | 9632134 |
| CDKN1B | CDKN4 | KIP1 | MEN1B | MEN4 | P27KIP1 | cyclin-dependent kinase inhibitor 1B (p27, Kip1) | - | HPRD | 7478582 |10908655 |
| CDKN1C | BWCR | BWS | KIP2 | WBS | p57 | cyclin-dependent kinase inhibitor 1C (p57, Kip2) | Reconstituted Complex | BioGRID | 10764802 |
| CDKN2A | ARF | CDK4I | CDKN2 | CMM2 | INK4 | INK4a | MLM | MTS1 | TP16 | p14 | p14ARF | p16 | p16INK4 | p16INK4a | p19 | cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | Reconstituted Complex | BioGRID | 10580009 |
| CDKN2D | INK4D | p19 | p19-INK4D | cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) | - | HPRD | 7739547 |
| CHUK | IKBKA | IKK-alpha | IKK1 | IKKA | NFKBIKA | TCF16 | conserved helix-loop-helix ubiquitous kinase | IKK-alpha interacts with cyclin D1 promoter. | BIND | 15808510 |
| CREB1 | CREB | MGC9284 | cAMP responsive element binding protein 1 | CREB interacts with CRE. | BIND | 15897899 |
| CREBBP | CBP | KAT3A | RSTS | CREB binding protein | CBP interacts with cyclin D1 promoter. | BIND | 15808510 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | CTNNB1 (beta-catenin) interacts with the CCND1 promoter. | BIND | 15592430 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | - | HPRD | 11836555 |
| CUL3 | - | cullin 3 | - | HPRD | 10500095 |
| DMTF1 | DMP1 | DMTF | FLJ41265 | hDMP1 | cyclin D binding myb-like transcription factor 1 | - | HPRD | 8887674 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | An unspecified isoform of EGFR binds to Cyclin D1 promoter. | BIND | 15950906 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | - | HPRD,BioGRID | 11788592 |
| ERCC3 | BTF2 | GTF2H | RAD25 | TFIIH | XPB | excision repair cross-complementing rodent repair deficiency, complementation group 3 (xeroderma pigmentosum group B complementing) | TFIIH interacts with Cyclin D1 promoter. | BIND | 15893730 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | - | HPRD,BioGRID | 9039267 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | ER-alpha interacts with cyclin D1 promoter. | BIND | 15808510 |
| FOXO4 | AFX | AFX1 | MGC120490 | MLLT7 | forkhead box O4 | MLLT7 (FOXO4) interacts with the CCND1 (Cyclin D1) promoter. This interaction was modelled on a demonstrated interaction between MLLT7 and CCND1 both from unspecified species. | BIND | 15688030 |
| HDAC3 | HD3 | RPD3 | RPD3-2 | histone deacetylase 3 | Affinity Capture-Western | BioGRID | 12048199 |15558026 |
| HERC5 | CEB1 | CEBP1 | hect domain and RLD 5 | - | HPRD,BioGRID | 10581175 |
| IFI27 | FAM14D | ISG12 | ISG12A | P27 | interferon, alpha-inducible protein 27 | - | HPRD | 9837900 |
| KAT2B | CAF | P | P/CAF | PCAF | K(lysine) acetyltransferase 2B | - | HPRD,BioGRID | 10318892 |
| KLF6 | BCD1 | COPEB | CPBP | DKFZp686N0199 | GBF | PAC1 | ST12 | ZF9 | Kruppel-like factor 6 | KLF6 interacts with Cyclin D1. | BIND | 15172998 |
| MCM10 | CNA43 | DNA43 | MGC126776 | PRO2249 | minichromosome maintenance complex component 10 | - | HPRD | 15232106 |
| MYBL2 | B-MYB | BMYB | MGC15600 | v-myb myeloblastosis viral oncogene homolog (avian)-like 2 | - | HPRD | 10645009 |
| NCOA1 | F-SRC-1 | KAT13A | MGC129719 | MGC129720 | NCoA-1 | RIP160 | SRC-1 | SRC1 | bHLHe42 | nuclear receptor coactivator 1 | - | HPRD,BioGRID | 9832502 |
| NCOA3 | ACTR | AIB-1 | AIB1 | CAGH16 | CTG26 | KAT13B | MGC141848 | RAC3 | SRC3 | TNRC14 | TNRC16 | TRAM-1 | pCIP | nuclear receptor coactivator 3 | - | HPRD,BioGRID | 9832502 |
| NCOA3 | ACTR | AIB-1 | AIB1 | CAGH16 | CTG26 | KAT13B | MGC141848 | RAC3 | SRC3 | TNRC14 | TNRC16 | TRAM-1 | pCIP | nuclear receptor coactivator 3 | AIB1 interacts with cyclin D1 promoter. | BIND | 15808510 |
| NEUROD1 | BETA2 | BHF-1 | NEUROD | bHLHa3 | neurogenic differentiation 1 | Affinity Capture-Western Phenotypic Suppression | BioGRID | 11788592 |
| NPDC1 | CAB | CAB- | CAB-1 | CAB1 | DKFZp586J0523 | neural proliferation, differentiation and control, 1 | - | HPRD,BioGRID | 11042687 |
| PCNA | MGC8367 | proliferating cell nuclear antigen | Reconstituted Complex | BioGRID | 7908906 |
| PIN1 | DOD | UBL5 | peptidylprolyl cis/trans isomerase, NIMA-interacting 1 | PIN1 interacts with Cyclin D1. This interaction was modelled on a demonstrated interaction between human PIN1 and Cyclin D1 from an unspecified species. | BIND | 11805292 |
| POLR2A | MGC75453 | POLR2 | POLRA | RPB1 | RPBh1 | RPO2 | RPOL2 | RpIILS | hRPB220 | hsRPB1 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | Pol II interacts with cyclin D1 promoter. | BIND | 15808510 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | Rb interacts with Cyclin D1. This interaction was modelled on a demonstrated interaction between Cyclin D1 from mouse and Rb from human. | BIND | 11126356 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | - | HPRD,BioGRID | 8490963 |
| RBL1 | CP107 | MGC40006 | PRB1 | p107 | retinoblastoma-like 1 (p107) | - | HPRD,BioGRID | 8643455 |
| RBX1 | BA554C12.1 | MGC13357 | MGC1481 | RNF75 | ROC1 | ring-box 1 | - | HPRD,BioGRID | 11311237 |
| RELA | MGC131774 | NFKB3 | p65 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | RELA (NF-kappaB p65 subunit) interacts with the CCND1 (Cyclin D1) promoter. | BIND | 15652748 |
| RFC1 | A1 | MGC51786 | MHCBFB | PO-GA | RECC1 | RFC | RFC140 | replication factor C (activator 1) 1, 145kDa | - | HPRD,BioGRID | 10353443 |
| STAT3 | APRF | FLJ20882 | HIES | MGC16063 | signal transducer and activator of transcription 3 (acute-phase response factor) | An unspecified isoform of STAT3 binds to Cyclin D1 promoter. | BIND | 15950906 |
| STAT3 | APRF | FLJ20882 | HIES | MGC16063 | signal transducer and activator of transcription 3 (acute-phase response factor) | - | HPRD,BioGRID | 11279133 |
| TAF1 | BA2R | CCG1 | CCGS | DYT3 | KAT4 | N-TAF1 | NSCL2 | OF | P250 | TAF2A | TAFII250 | TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa | TAFII250 interacts with Cyclin D1. This interaction was modelled on a demonstrated interaction between TAFII250 from human and Cyclin D1 from mouse. | BIND | 11126356 |
| TAF1 | BA2R | CCG1 | CCGS | DYT3 | KAT4 | N-TAF1 | NSCL2 | OF | P250 | TAF2A | TAFII250 | TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa | - | HPRD,BioGRID | 9926939 |11126356 |
| THRA | AR7 | EAR7 | ERB-T-1 | ERBA | ERBA1 | MGC000261 | MGC43240 | NR1A1 | THRA1 | THRA2 | c-ERBA-1 | thyroid hormone receptor, alpha (erythroblastic leukemia viral (v-erb-a) oncogene homolog, avian) | - | HPRD,BioGRID | 12048199 |
| THRB | ERBA-BETA | ERBA2 | GRTH | MGC126109 | MGC126110 | NR1A2 | PRTH | THR1 | THRB1 | THRB2 | thyroid hormone receptor, beta (erythroblastic leukemia viral (v-erb-a) oncogene homolog 2, avian) | Affinity Capture-Western Reconstituted Complex | BioGRID | 12048199 |
| TSC2 | FLJ43106 | LAM | TSC4 | tuberous sclerosis 2 | Affinity Capture-Western | BioGRID | 16357142 |
| XPO1 | CRM1 | DKFZp686B1823 | exportin 1 (CRM1 homolog, yeast) | Reconstituted Complex | BioGRID | 12529437 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CELL CYCLE | 128 | 84 | All SZGR 2.0 genes in this pathway |
| KEGG P53 SIGNALING PATHWAY | 69 | 45 | All SZGR 2.0 genes in this pathway |
| KEGG WNT SIGNALING PATHWAY | 151 | 112 | All SZGR 2.0 genes in this pathway |
| KEGG FOCAL ADHESION | 201 | 138 | All SZGR 2.0 genes in this pathway |
| KEGG JAK STAT SIGNALING PATHWAY | 155 | 105 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG COLORECTAL CANCER | 62 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG PANCREATIC CANCER | 70 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG ENDOMETRIAL CANCER | 52 | 45 | All SZGR 2.0 genes in this pathway |
| KEGG GLIOMA | 65 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG PROSTATE CANCER | 89 | 75 | All SZGR 2.0 genes in this pathway |
| KEGG THYROID CANCER | 29 | 26 | All SZGR 2.0 genes in this pathway |
| KEGG MELANOMA | 71 | 57 | All SZGR 2.0 genes in this pathway |
| KEGG BLADDER CANCER | 42 | 33 | All SZGR 2.0 genes in this pathway |
| KEGG CHRONIC MYELOID LEUKEMIA | 73 | 59 | All SZGR 2.0 genes in this pathway |
| KEGG ACUTE MYELOID LEUKEMIA | 60 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG SMALL CELL LUNG CANCER | 84 | 67 | All SZGR 2.0 genes in this pathway |
| KEGG NON SMALL CELL LUNG CANCER | 54 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG VIRAL MYOCARDITIS | 73 | 58 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CARM ER PATHWAY | 35 | 27 | All SZGR 2.0 genes in this pathway |
| BIOCARTA G1 PATHWAY | 28 | 21 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CELLCYCLE PATHWAY | 23 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GSK3 PATHWAY | 27 | 26 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RACCYCD PATHWAY | 26 | 23 | All SZGR 2.0 genes in this pathway |
| BIOCARTA P53 PATHWAY | 16 | 13 | All SZGR 2.0 genes in this pathway |
| BIOCARTA WNT PATHWAY | 26 | 24 | All SZGR 2.0 genes in this pathway |
| WNT SIGNALING | 89 | 71 | All SZGR 2.0 genes in this pathway |
| SA G1 AND S PHASES | 15 | 10 | All SZGR 2.0 genes in this pathway |
| SA REG CASCADE OF CYCLIN EXPR | 13 | 7 | All SZGR 2.0 genes in this pathway |
| PID NOTCH PATHWAY | 59 | 49 | All SZGR 2.0 genes in this pathway |
| PID AR PATHWAY | 61 | 46 | All SZGR 2.0 genes in this pathway |
| PID FRA PATHWAY | 37 | 28 | All SZGR 2.0 genes in this pathway |
| PID PS1 PATHWAY | 46 | 39 | All SZGR 2.0 genes in this pathway |
| PID ILK PATHWAY | 45 | 32 | All SZGR 2.0 genes in this pathway |
| PID TELOMERASE PATHWAY | 68 | 48 | All SZGR 2.0 genes in this pathway |
| PID ECADHERIN NASCENT AJ PATHWAY | 39 | 33 | All SZGR 2.0 genes in this pathway |
| PID ATF2 PATHWAY | 59 | 43 | All SZGR 2.0 genes in this pathway |
| PID AP1 PATHWAY | 70 | 60 | All SZGR 2.0 genes in this pathway |
| PID FOXM1 PATHWAY | 40 | 30 | All SZGR 2.0 genes in this pathway |
| PID TRKR PATHWAY | 62 | 48 | All SZGR 2.0 genes in this pathway |
| PID CMYB PATHWAY | 84 | 61 | All SZGR 2.0 genes in this pathway |
| PID ERA GENOMIC PATHWAY | 65 | 37 | All SZGR 2.0 genes in this pathway |
| PID P38 GAMMA DELTA PATHWAY | 11 | 9 | All SZGR 2.0 genes in this pathway |
| PID BETA CATENIN NUC PATHWAY | 80 | 60 | All SZGR 2.0 genes in this pathway |
| PID MYC REPRESS PATHWAY | 63 | 52 | All SZGR 2.0 genes in this pathway |
| PID PI3K PLC TRK PATHWAY | 36 | 31 | All SZGR 2.0 genes in this pathway |
| PID RB 1PATHWAY | 65 | 46 | All SZGR 2.0 genes in this pathway |
| PID FAK PATHWAY | 59 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | 29 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME PRE NOTCH EXPRESSION AND PROCESSING | 44 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME G1 PHASE | 38 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC G1 G1 S PHASES | 137 | 79 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH | 103 | 64 | All SZGR 2.0 genes in this pathway |
| REACTOME S PHASE | 109 | 66 | All SZGR 2.0 genes in this pathway |
| PYEON HPV POSITIVE TUMORS DN | 10 | 5 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| KOBAYASHI EGFR SIGNALING 6HR DN | 18 | 13 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS DN | 182 | 111 | All SZGR 2.0 genes in this pathway |
| WILCOX RESPONSE TO PROGESTERONE DN | 66 | 44 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL UP | 380 | 215 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL UP | 133 | 78 | All SZGR 2.0 genes in this pathway |
| NOJIMA SFRP2 TARGETS DN | 25 | 18 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL DN | 275 | 168 | All SZGR 2.0 genes in this pathway |
| BOGNI TREATMENT RELATED MYELOID LEUKEMIA DN | 33 | 19 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC DN | 187 | 115 | All SZGR 2.0 genes in this pathway |
| LOPEZ MESOTHELIOMA SURVIVAL WORST VS BEST UP | 14 | 10 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN | 459 | 276 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 12HR DN | 209 | 122 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP UP | 265 | 158 | All SZGR 2.0 genes in this pathway |
| PROVENZANI METASTASIS DN | 136 | 94 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION KRAS CDC25 UP | 58 | 39 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION KRAS UP | 126 | 72 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 DN | 229 | 142 | All SZGR 2.0 genes in this pathway |
| CHIN BREAST CANCER COPY NUMBER UP | 27 | 18 | All SZGR 2.0 genes in this pathway |
| VANHARANTA UTERINE FIBROID UP | 45 | 26 | All SZGR 2.0 genes in this pathway |
| CAIRO PML TARGETS BOUND BY MYC DN | 14 | 12 | All SZGR 2.0 genes in this pathway |
| LANDIS ERBB2 BREAST TUMORS 324 UP | 150 | 93 | All SZGR 2.0 genes in this pathway |
| BARIS THYROID CANCER DN | 59 | 45 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| JOHANSSON GLIOMAGENESIS BY PDGFB UP | 58 | 44 | All SZGR 2.0 genes in this pathway |
| LUI THYROID CANCER PAX8 PPARG UP | 44 | 29 | All SZGR 2.0 genes in this pathway |
| HUMMERICH MALIGNANT SKIN TUMOR UP | 16 | 8 | All SZGR 2.0 genes in this pathway |
| BUSA SAM68 TARGETS UP | 7 | 6 | All SZGR 2.0 genes in this pathway |
| MARKS HDAC TARGETS DN | 15 | 11 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 TTD DN | 84 | 63 | All SZGR 2.0 genes in this pathway |
| BAKER HEMATOPOIESIS STAT3 TARGETS | 16 | 12 | All SZGR 2.0 genes in this pathway |
| BAKER HEMATOPOESIS STAT5 TARGETS | 7 | 7 | All SZGR 2.0 genes in this pathway |
| SANCHEZ MDM2 TARGETS | 15 | 10 | All SZGR 2.0 genes in this pathway |
| SCHEIDEREIT IKK TARGETS | 18 | 15 | All SZGR 2.0 genes in this pathway |
| SUZUKI AMPLIFIED IN ORAL CANCER | 16 | 12 | All SZGR 2.0 genes in this pathway |
| MYLLYKANGAS AMPLIFICATION HOT SPOT 23 | 24 | 13 | All SZGR 2.0 genes in this pathway |
| LUI THYROID CANCER CLUSTER 1 | 51 | 33 | All SZGR 2.0 genes in this pathway |
| MATTIOLI MULTIPLE MYELOMA WITH 14Q32 TRANSLOCATIONS | 36 | 25 | All SZGR 2.0 genes in this pathway |
| SCHEIDEREIT IKK INTERACTING PROTEINS | 58 | 45 | All SZGR 2.0 genes in this pathway |
| HONRADO BREAST CANCER BRCA1 VS BRCA2 | 18 | 12 | All SZGR 2.0 genes in this pathway |
| SNIJDERS AMPLIFIED IN HEAD AND NECK TUMORS | 37 | 27 | All SZGR 2.0 genes in this pathway |
| SCHRAMM INHBA TARGETS UP | 7 | 7 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS NEUROEPITHELIUM UP | 176 | 111 | All SZGR 2.0 genes in this pathway |
| BORLAK LIVER CANCER EGF UP | 57 | 41 | All SZGR 2.0 genes in this pathway |
| OXFORD RALA OR RALB TARGETS UP | 48 | 23 | All SZGR 2.0 genes in this pathway |
| MAINA VHL TARGETS DN | 18 | 14 | All SZGR 2.0 genes in this pathway |
| SIMBULAN UV RESPONSE IMMORTALIZED DN | 31 | 26 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM2 | 153 | 102 | All SZGR 2.0 genes in this pathway |
| PUJANA BREAST CANCER LIT INT NETWORK | 101 | 73 | All SZGR 2.0 genes in this pathway |
| OHM METHYLATED IN ADULT CANCERS | 27 | 23 | All SZGR 2.0 genes in this pathway |
| GROSS ELK3 TARGETS UP | 27 | 16 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 UP | 209 | 139 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 AND HIF1A DN | 103 | 71 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| SCHWAB TARGETS OF BMYB POLYMORPHIC VARIANTS UP | 13 | 9 | All SZGR 2.0 genes in this pathway |
| FRIDMAN SENESCENCE UP | 77 | 60 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 60 HELA | 46 | 32 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 480 HELA | 164 | 118 | All SZGR 2.0 genes in this pathway |
| SHIN B CELL LYMPHOMA CLUSTER 3 | 28 | 23 | All SZGR 2.0 genes in this pathway |
| LEE TARGETS OF PTCH1 AND SUFU UP | 53 | 37 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 11Q12 Q14 AMPLICON | 158 | 93 | All SZGR 2.0 genes in this pathway |
| STANELLE E2F1 TARGETS | 29 | 20 | All SZGR 2.0 genes in this pathway |
| CHESLER BRAIN QTL CIS | 75 | 51 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC TGFA UP | 61 | 40 | All SZGR 2.0 genes in this pathway |
| WILLERT WNT SIGNALING | 24 | 13 | All SZGR 2.0 genes in this pathway |
| LIN APC TARGETS | 77 | 55 | All SZGR 2.0 genes in this pathway |
| LE EGR2 TARGETS UP | 108 | 75 | All SZGR 2.0 genes in this pathway |
| BECKER TAMOXIFEN RESISTANCE DN | 52 | 37 | All SZGR 2.0 genes in this pathway |
| PENG GLUTAMINE DEPRIVATION DN | 337 | 230 | All SZGR 2.0 genes in this pathway |
| ROSS AML WITH AML1 ETO FUSION | 76 | 55 | All SZGR 2.0 genes in this pathway |
| ZHANG TARGETS OF EWSR1 FLI1 FUSION | 88 | 68 | All SZGR 2.0 genes in this pathway |
| VERNELL RETINOBLASTOMA PATHWAY DN | 22 | 13 | All SZGR 2.0 genes in this pathway |
| HOFMANN CELL LYMPHOMA UP | 50 | 35 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA SPIKED | 22 | 13 | All SZGR 2.0 genes in this pathway |
| LEI MYB TARGETS | 318 | 215 | All SZGR 2.0 genes in this pathway |
| ABRAHAM ALPC VS MULTIPLE MYELOMA UP | 26 | 22 | All SZGR 2.0 genes in this pathway |
| STAEGE EWING FAMILY TUMOR | 33 | 22 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| RAMALHO STEMNESS UP | 206 | 118 | All SZGR 2.0 genes in this pathway |
| LEE AGING CEREBELLUM DN | 86 | 66 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY UP | 86 | 48 | All SZGR 2.0 genes in this pathway |
| SIMBULAN PARP1 TARGETS UP | 31 | 23 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| ZHANG PROLIFERATING VS QUIESCENT | 51 | 41 | All SZGR 2.0 genes in this pathway |
| GERHOLD ADIPOGENESIS DN | 64 | 44 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 24 HR DN | 91 | 64 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL DN | 128 | 93 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 7 | 51 | 34 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER BRCA1 VS BRCA2 | 163 | 113 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS DN | 141 | 92 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| SANSOM APC MYC TARGETS | 217 | 138 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY GAMMA AND UV RADIATION | 88 | 65 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE DN | 195 | 135 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS DN | 292 | 189 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION DN | 281 | 179 | All SZGR 2.0 genes in this pathway |
| AMUNDSON POOR SURVIVAL AFTER GAMMA RADIATION 8G | 95 | 62 | All SZGR 2.0 genes in this pathway |
| AMUNDSON POOR SURVIVAL AFTER GAMMA RADIATION 2G | 171 | 96 | All SZGR 2.0 genes in this pathway |
| LIN MELANOMA COPY NUMBER UP | 73 | 53 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| WALLACE PROSTATE CANCER RACE UP | 299 | 167 | All SZGR 2.0 genes in this pathway |
| MARZEC IL2 SIGNALING UP | 115 | 80 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE DN | 274 | 165 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B UP | 172 | 109 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| BASSO HAIRY CELL LEUKEMIA DN | 80 | 66 | All SZGR 2.0 genes in this pathway |
| ZHENG GLIOBLASTOMA PLASTICITY UP | 250 | 168 | All SZGR 2.0 genes in this pathway |
| BONCI TARGETS OF MIR15A AND MIR16 1 | 91 | 75 | All SZGR 2.0 genes in this pathway |
| LABBE WNT3A TARGETS UP | 112 | 71 | All SZGR 2.0 genes in this pathway |
| CLIMENT BREAST CANCER COPY NUMBER UP | 23 | 20 | All SZGR 2.0 genes in this pathway |
| BREDEMEYER RAG SIGNALING NOT VIA ATM UP | 62 | 29 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| BEIER GLIOMA STEM CELL DN | 66 | 42 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA D CLUSTER UP | 27 | 19 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA D UP | 89 | 62 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP | 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| SWEET KRAS ONCOGENIC SIGNATURE | 89 | 56 | All SZGR 2.0 genes in this pathway |
| HOFMANN MYELODYSPLASTIC SYNDROM LOW RISK DN | 30 | 20 | All SZGR 2.0 genes in this pathway |
| HOFMANN MYELODYSPLASTIC SYNDROM RISK UP | 24 | 14 | All SZGR 2.0 genes in this pathway |
| COLINA TARGETS OF 4EBP1 AND 4EBP2 | 356 | 214 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN PARTIALLY REPROGRAMMED TO PLURIPOTENCY | 10 | 6 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| LOPEZ MESOTHELIOMA SURVIVAL OVERALL DN | 16 | 8 | All SZGR 2.0 genes in this pathway |
| KOBAYASHI EGFR SIGNALING 24HR DN | 251 | 151 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA PCA3 UP | 80 | 54 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN | 253 | 192 | All SZGR 2.0 genes in this pathway |
| DORN ADENOVIRUS INFECTION 12HR DN | 33 | 25 | All SZGR 2.0 genes in this pathway |
| DORN ADENOVIRUS INFECTION 24HR DN | 43 | 32 | All SZGR 2.0 genes in this pathway |
| DORN ADENOVIRUS INFECTION 32HR DN | 39 | 28 | All SZGR 2.0 genes in this pathway |
| DORN ADENOVIRUS INFECTION 48HR DN | 40 | 29 | All SZGR 2.0 genes in this pathway |
| WONG EMBRYONIC STEM CELL CORE | 335 | 193 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 13 | 172 | 107 | All SZGR 2.0 genes in this pathway |
| SUBTIL PROGESTIN TARGETS | 36 | 25 | All SZGR 2.0 genes in this pathway |
| CARD MIR302A TARGETS | 77 | 62 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS DN | 213 | 127 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS NOT VIA AKT1 UP | 211 | 131 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS VIA AKT1 UP | 281 | 183 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| IVANOVSKA MIR106B TARGETS | 90 | 56 | All SZGR 2.0 genes in this pathway |
| WIEDERSCHAIN TARGETS OF BMI1 AND PCGF2 | 57 | 39 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION DN | 321 | 200 | All SZGR 2.0 genes in this pathway |
| WACKER HYPOXIA TARGETS OF VHL | 13 | 11 | All SZGR 2.0 genes in this pathway |
| DELACROIX RARG BOUND MEF | 367 | 231 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR TARGETS DN | 24 | 16 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR BOUND ES | 462 | 273 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| BOUDOUKHA BOUND BY IGF2BP2 | 111 | 59 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| let-7/98 | 2214 | 2220 | 1A | hsa-let-7abrain | UGAGGUAGUAGGUUGUAUAGUU |
| hsa-let-7bbrain | UGAGGUAGUAGGUUGUGUGGUU | ||||
| hsa-let-7cbrain | UGAGGUAGUAGGUUGUAUGGUU | ||||
| hsa-let-7dbrain | AGAGGUAGUAGGUUGCAUAGU | ||||
| hsa-let-7ebrain | UGAGGUAGGAGGUUGUAUAGU | ||||
| hsa-let-7fbrain | UGAGGUAGUAGAUUGUAUAGUU | ||||
| hsa-miR-98brain | UGAGGUAGUAAGUUGUAUUGUU | ||||
| hsa-let-7gSZ | UGAGGUAGUAGUUUGUACAGU | ||||
| hsa-let-7ibrain | UGAGGUAGUAGUUUGUGCUGU | ||||
| miR-1/206 | 1000 | 1006 | 1A | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-15/16/195/424/497 | 2033 | 2040 | 1A,m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG | ||||
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-17-5p/20/93.mr/106/519.d | 1014 | 1020 | m8 | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-19 | 1778 | 1785 | 1A,m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-23 | 2171 | 2177 | m8 | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-323 | 2170 | 2177 | 1A,m8 | hsa-miR-323brain | GCACAUUACACGGUCGACCUCU |
| miR-338 | 907 | 913 | m8 | hsa-miR-338brain | UCCAGCAUCAGUGAUUUUGUUGA |
| miR-34/449 | 2076 | 2082 | 1A | hsa-miR-34abrain | UGGCAGUGUCUUAGCUGGUUGUU |
| hsa-miR-34c | AGGCAGUGUAGUUAGCUGAUUGC | ||||
| hsa-miR-449 | UGGCAGUGUAUUGUUAGCUGGU | ||||
| hsa-miR-449b | AGGCAGUGUAUUGUUAGCUGGC | ||||
| miR-503 | 1960 | 1967 | 1A,m8 | hsa-miR-503 | UAGCAGCGGGAACAGUUCUGCAG |
| hsa-miR-503 | UAGCAGCGGGAACAGUUCUGCAG | ||||
| miR-93.hd/291-3p/294/295/302/372/373/520 | 1013 | 1019 | m8 | hsa-miR-93brain | AAAGUGCUGUUCGUGCAGGUAG |
| hsa-miR-302a | UAAGUGCUUCCAUGUUUUGGUGA | ||||
| hsa-miR-302b | UAAGUGCUUCCAUGUUUUAGUAG | ||||
| hsa-miR-302c | UAAGUGCUUCCAUGUUUCAGUGG | ||||
| hsa-miR-302d | UAAGUGCUUCCAUGUUUGAGUGU | ||||
| hsa-miR-372 | AAAGUGCUGCGACAUUUGAGCGU | ||||
| hsa-miR-373 | GAAGUGCUUCGAUUUUGGGGUGU | ||||
| hsa-miR-520e | AAAGUGCUUCCUUUUUGAGGG | ||||
| hsa-miR-520a | AAAGUGCUUCCCUUUGGACUGU | ||||
| hsa-miR-520b | AAAGUGCUUCCUUUUAGAGGG | ||||
| hsa-miR-520c | AAAGUGCUUCCUUUUAGAGGGUU | ||||
| hsa-miR-520d | AAAGUGCUUCUCUUUGGUGGGUU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.