Gene Page: CAST
Summary ?
| GeneID | 831 |
| Symbol | CAST |
| Synonyms | BS-17|PLACK |
| Description | calpastatin |
| Reference | MIM:114090|HGNC:HGNC:1515|Ensembl:ENSG00000153113|HPRD:00233|Vega:OTTHUMG00000128413 |
| Gene type | protein-coding |
| Map location | 5q15 |
| Pascal p-value | 0.246 |
| Fetal beta | -1.808 |
| eGene | Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Frontal Cortex BA9 Hippocampus Nucleus accumbens basal ganglia Putamen basal ganglia Myers' cis & trans Meta |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizotypy,schizophrenias,schizotypal | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6046959 | chr20 | 20588433 | CAST | 831 | 0.07 | trans | ||
| rs2144280 | chr20 | 20655465 | CAST | 831 | 0.19 | trans | ||
| rs10075736 | 5 | 95990057 | CAST | ENSG00000153113.19 | 5.181E-9 | 0 | 129086 | gtex_brain_putamen_basal |
| rs10039522 | 5 | 95990869 | CAST | ENSG00000153113.19 | 5.187E-9 | 0 | 129898 | gtex_brain_putamen_basal |
| rs10045215 | 5 | 95990871 | CAST | ENSG00000153113.19 | 5.187E-9 | 0 | 129900 | gtex_brain_putamen_basal |
| rs2351245 | 5 | 95995733 | CAST | ENSG00000153113.19 | 6.734E-12 | 0 | 134762 | gtex_brain_putamen_basal |
| rs4267886 | 5 | 95997266 | CAST | ENSG00000153113.19 | 6.734E-12 | 0 | 136295 | gtex_brain_putamen_basal |
| rs4590196 | 5 | 95997285 | CAST | ENSG00000153113.19 | 2.045E-10 | 0 | 136314 | gtex_brain_putamen_basal |
| rs3048781 | 5 | 95997566 | CAST | ENSG00000153113.19 | 7.901E-10 | 0 | 136595 | gtex_brain_putamen_basal |
| rs200953056 | 5 | 95997577 | CAST | ENSG00000153113.19 | 1.746E-6 | 0 | 136606 | gtex_brain_putamen_basal |
| rs7733671 | 5 | 96000269 | CAST | ENSG00000153113.19 | 2.048E-10 | 0 | 139298 | gtex_brain_putamen_basal |
| rs6860934 | 5 | 96001842 | CAST | ENSG00000153113.19 | 1.764E-10 | 0 | 140871 | gtex_brain_putamen_basal |
| rs10067766 | 5 | 96002020 | CAST | ENSG00000153113.19 | 1.757E-10 | 0 | 141049 | gtex_brain_putamen_basal |
| rs6897588 | 5 | 96003149 | CAST | ENSG00000153113.19 | 2.449E-10 | 0 | 142178 | gtex_brain_putamen_basal |
| rs7704167 | 5 | 96004363 | CAST | ENSG00000153113.19 | 9.818E-12 | 0 | 143392 | gtex_brain_putamen_basal |
| rs7705824 | 5 | 96005617 | CAST | ENSG00000153113.19 | 2.747E-10 | 0 | 144646 | gtex_brain_putamen_basal |
| rs31250 | 5 | 96013441 | CAST | ENSG00000153113.19 | 2.496E-10 | 0 | 152470 | gtex_brain_putamen_basal |
| rs31249 | 5 | 96013931 | CAST | ENSG00000153113.19 | 9.818E-12 | 0 | 152960 | gtex_brain_putamen_basal |
| rs31248 | 5 | 96014683 | CAST | ENSG00000153113.19 | 2.842E-10 | 0 | 153712 | gtex_brain_putamen_basal |
| rs26485 | 5 | 96016477 | CAST | ENSG00000153113.19 | 2.58E-10 | 0 | 155506 | gtex_brain_putamen_basal |
| rs26484 | 5 | 96016569 | CAST | ENSG00000153113.19 | 1.667E-8 | 0 | 155598 | gtex_brain_putamen_basal |
| rs26483 | 5 | 96016616 | CAST | ENSG00000153113.19 | 5.022E-10 | 0 | 155645 | gtex_brain_putamen_basal |
| rs34761761 | 5 | 96017666 | CAST | ENSG00000153113.19 | 2.105E-9 | 0 | 156695 | gtex_brain_putamen_basal |
| rs26482 | 5 | 96017956 | CAST | ENSG00000153113.19 | 2.09E-8 | 0 | 156985 | gtex_brain_putamen_basal |
| rs28040 | 5 | 96018066 | CAST | ENSG00000153113.19 | 2.167E-8 | 0 | 157095 | gtex_brain_putamen_basal |
| rs152005 | 5 | 96025847 | CAST | ENSG00000153113.19 | 2.21E-7 | 0 | 164876 | gtex_brain_putamen_basal |
| rs152020 | 5 | 96026085 | CAST | ENSG00000153113.19 | 5.273E-10 | 0 | 165114 | gtex_brain_putamen_basal |
| rs152014 | 5 | 96027252 | CAST | ENSG00000153113.19 | 1.264E-9 | 0 | 166281 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
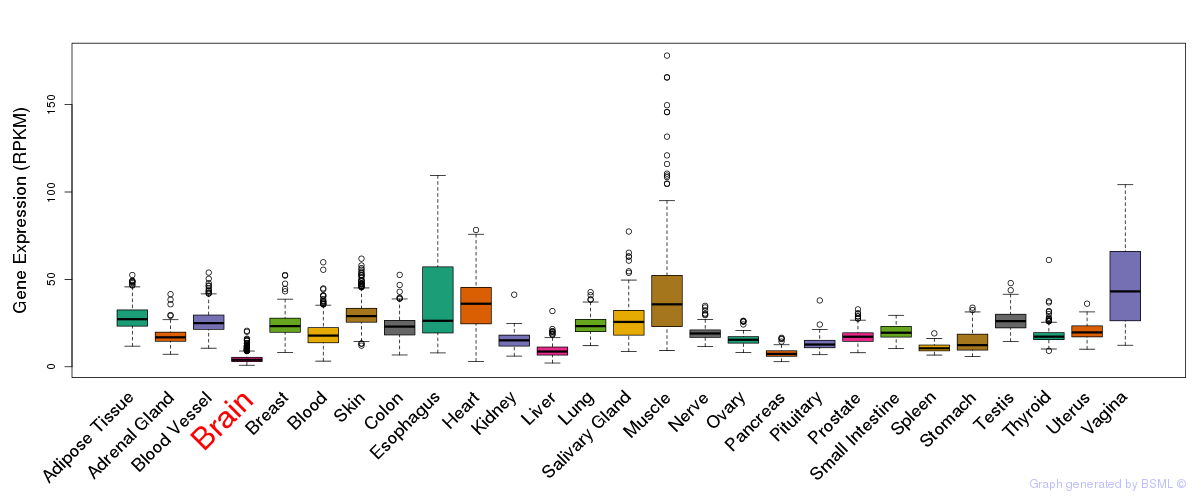
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PHF2 | 0.97 | 0.94 |
| CCNJ | 0.97 | 0.93 |
| KIAA1267 | 0.96 | 0.96 |
| ZNF532 | 0.96 | 0.95 |
| SMARCD1 | 0.96 | 0.95 |
| BRD1 | 0.96 | 0.95 |
| BCL9 | 0.96 | 0.94 |
| C11orf30 | 0.96 | 0.95 |
| CNOT6 | 0.96 | 0.94 |
| NUP188 | 0.95 | 0.93 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.72 | -0.80 |
| AIFM3 | -0.70 | -0.77 |
| HLA-F | -0.69 | -0.74 |
| FBXO2 | -0.68 | -0.65 |
| ALDOC | -0.68 | -0.70 |
| S100B | -0.68 | -0.84 |
| PTH1R | -0.68 | -0.72 |
| AF347015.31 | -0.68 | -0.89 |
| LHPP | -0.67 | -0.60 |
| AF347015.27 | -0.67 | -0.87 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN | 463 | 290 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LPS UP | 431 | 237 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 UP | 121 | 71 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS F UP | 185 | 119 | All SZGR 2.0 genes in this pathway |
| WANG ESOPHAGUS CANCER VS NORMAL DN | 101 | 66 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| MEINHOLD OVARIAN CANCER LOW GRADE UP | 19 | 15 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO BEXAROTENE DN | 29 | 19 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR UP | 176 | 115 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT AND CANCER BOX2 DN | 7 | 6 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT AND CANCER BOX5 DN | 8 | 5 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS UP | 424 | 268 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED UP | 183 | 111 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 1 UP | 182 | 119 | All SZGR 2.0 genes in this pathway |
| YAGI AML FAB MARKERS | 191 | 131 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT UP | 390 | 242 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS MATURE CELL | 293 | 160 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| YIH RESPONSE TO ARSENITE C2 | 18 | 15 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN UP | 262 | 186 | All SZGR 2.0 genes in this pathway |
| TAKAO RESPONSE TO UVB RADIATION DN | 98 | 67 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| DURCHDEWALD SKIN CARCINOGENESIS DN | 264 | 168 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE DN | 373 | 196 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER T7 | 98 | 63 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| ZHENG FOXP3 TARGETS IN THYMUS UP | 196 | 137 | All SZGR 2.0 genes in this pathway |
| STEARMAN LUNG CANCER EARLY VS LATE UP | 125 | 89 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP | 578 | 341 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS DN | 145 | 93 | All SZGR 2.0 genes in this pathway |
| EHLERS ANEUPLOIDY UP | 41 | 29 | All SZGR 2.0 genes in this pathway |
| LABBE TGFB1 TARGETS UP | 102 | 64 | All SZGR 2.0 genes in this pathway |
| NAKAMURA METASTASIS MODEL DN | 43 | 28 | All SZGR 2.0 genes in this pathway |
| BHATI G2M ARREST BY 2METHOXYESTRADIOL DN | 127 | 75 | All SZGR 2.0 genes in this pathway |
| HUANG DASATINIB RESISTANCE UP | 81 | 53 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS UP | 165 | 118 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| WENDT COHESIN TARGETS UP | 33 | 19 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED | 351 | 238 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN | 253 | 192 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA MESENCHYMAL | 216 | 130 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR DN | 505 | 328 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| IKEDA MIR30 TARGETS UP | 116 | 87 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |