Gene Page: APOE
Summary ?
| GeneID | 348 |
| Symbol | APOE |
| Synonyms | AD2|APO-E|LDLCQ5|LPG |
| Description | apolipoprotein E |
| Reference | MIM:107741|HGNC:HGNC:613|Ensembl:ENSG00000130203|HPRD:00135|Vega:OTTHUMG00000128901 |
| Gene type | protein-coding |
| Map location | 19q13.2 |
| Pascal p-value | 0.019 |
| Sherlock p-value | 0.298 |
| Fetal beta | -2.818 |
| eGene | Myers' cis & trans |
| Support | CELL METABOLISM CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 2 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 5 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 31.2393 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16938766 | chr8 | 74949940 | APOE | 348 | 0.15 | trans | ||
| rs12545427 | chr8 | 74954879 | APOE | 348 | 0.04 | trans | ||
| rs6472816 | chr8 | 74961543 | APOE | 348 | 0.06 | trans | ||
| rs4783568 | chr16 | 66602031 | APOE | 348 | 0.05 | trans |
Section II. Transcriptome annotation
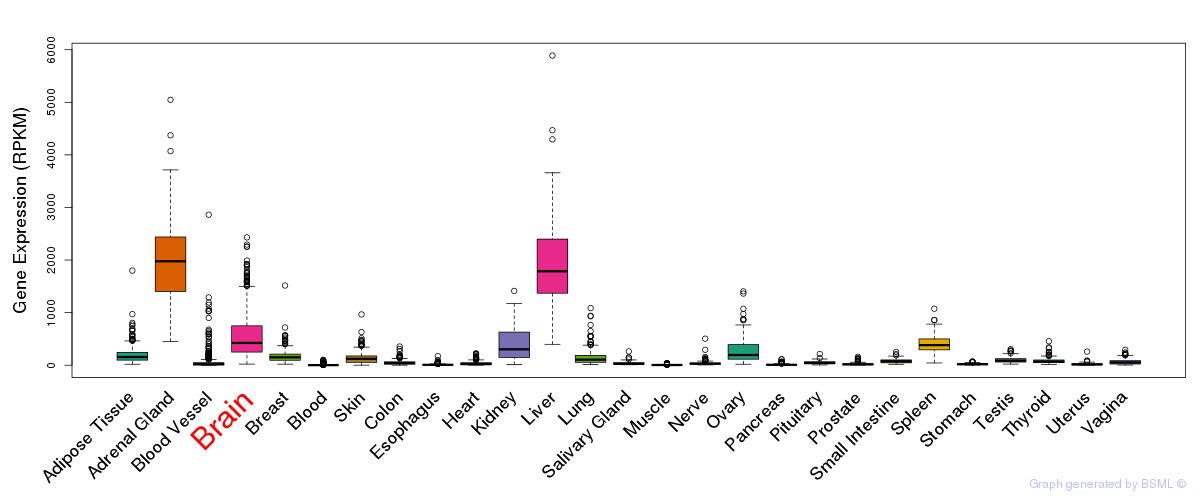
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| APLP2 | 0.93 | 0.92 |
| SV2A | 0.91 | 0.91 |
| MAGEE1 | 0.90 | 0.92 |
| DCTN1 | 0.90 | 0.90 |
| STXBP1 | 0.90 | 0.90 |
| WARS | 0.90 | 0.89 |
| MADD | 0.90 | 0.89 |
| DPP6 | 0.89 | 0.91 |
| NFE2L1 | 0.89 | 0.89 |
| CLSTN1 | 0.89 | 0.91 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.56 | -0.43 |
| C1orf61 | -0.56 | -0.61 |
| C1orf54 | -0.54 | -0.49 |
| GNG11 | -0.54 | -0.47 |
| AP002478.3 | -0.54 | -0.50 |
| HIGD1B | -0.51 | -0.42 |
| PLA2G5 | -0.50 | -0.38 |
| AF347015.31 | -0.50 | -0.42 |
| RAB34 | -0.50 | -0.53 |
| MT-CO2 | -0.50 | -0.42 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0001540 | beta-amyloid binding | IDA | 11305869 | |
| GO:0005543 | phospholipid binding | IDA | 4066713 | |
| GO:0005319 | lipid transporter activity | IEA | - | |
| GO:0008201 | heparin binding | IEA | - | |
| GO:0008289 | lipid binding | IEA | - | |
| GO:0008034 | lipoprotein binding | IEA | - | |
| GO:0017127 | cholesterol transporter activity | IEA | - | |
| GO:0016209 | antioxidant activity | IDA | 9685360 | |
| GO:0042803 | protein homodimerization activity | IDA | 8245722 | |
| GO:0046911 | metal chelating activity | IDA | 9685360 | |
| GO:0048156 | tau protein binding | IPI | 7566652 | |
| GO:0050749 | apolipoprotein E receptor binding | IDA | 1384047 | |
| GO:0050749 | apolipoprotein E receptor binding | IPI | 12950167 | |
| GO:0046982 | protein heterodimerization activity | IPI | 8245722 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007271 | synaptic transmission, cholinergic | TAS | neuron, Cholinergic, Synap, Neurotransmitter (GO term level: 7) | 9622609 |
| GO:0030516 | regulation of axon extension | TAS | axon (GO term level: 14) | 9622609 |
| GO:0048168 | regulation of neuronal synaptic plasticity | TAS | neuron, Synap (GO term level: 9) | 9622609 |
| GO:0000302 | response to reactive oxygen species | NAS | 11743999 | |
| GO:0001937 | negative regulation of endothelial cell proliferation | IDA | 9685360 | |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | IDA | 16443932 | |
| GO:0007263 | nitric oxide mediated signal transduction | IDA | 8995232 | |
| GO:0010544 | negative regulation of platelet activation | IDA | 8995232 | |
| GO:0006917 | induction of apoptosis | IDA | 12753088 | |
| GO:0007010 | cytoskeleton organization | TAS | 9622609 | |
| GO:0006707 | cholesterol catabolic process | IEA | - | |
| GO:0006979 | response to oxidative stress | IEA | - | |
| GO:0006641 | triacylglycerol metabolic process | IDA | 9649566 | |
| GO:0006641 | triacylglycerol metabolic process | IMP | 3771793 | |
| GO:0006916 | anti-apoptosis | IDA | 9685360 | |
| GO:0006869 | lipid transport | IEA | - | |
| GO:0006874 | cellular calcium ion homeostasis | IEA | - | |
| GO:0042311 | vasodilation | IEA | - | |
| GO:0033344 | cholesterol efflux | IEA | - | |
| GO:0051044 | positive regulation of membrane protein ectodomain proteolysis | IDA | 15950758 | |
| GO:0051000 | positive regulation of nitric-oxide synthase activity | IDA | 8995232 | |
| GO:0034447 | very-low-density lipoprotein particle clearance | IMP | 9649566 | |
| GO:0042158 | lipoprotein biosynthetic process | IEA | - | |
| GO:0019934 | cGMP-mediated signaling | IDA | 8995232 | |
| GO:0030828 | positive regulation of cGMP biosynthetic process | IDA | 8995232 | |
| GO:0032805 | positive regulation of low-density lipoprotein receptor catabolic process | IDA | 15950758 | |
| GO:0034382 | chylomicron remnant clearance | IMP | 7175379 | |
| GO:0032488 | Cdc42 protein signal transduction | IDA | 16443932 | |
| GO:0033700 | phospholipid efflux | IDA | 11162594 | |
| GO:0043407 | negative regulation of MAP kinase activity | IDA | 9685360 | |
| GO:0042632 | cholesterol homeostasis | IEA | - | |
| GO:0046907 | intracellular transport | TAS | 9622609 | |
| GO:0050728 | negative regulation of inflammatory response | IC | 8995232 | |
| GO:0043537 | negative regulation of blood vessel endothelial cell migration | IDA | 9685360 | |
| GO:0043691 | reverse cholesterol transport | IDA | 8127890 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0043025 | cell soma | NAS | axon, dendrite (GO term level: 4) | 8083695 |
| GO:0030425 | dendrite | NAS | neuron, axon, dendrite (GO term level: 6) | 8083695 |
| GO:0005576 | extracellular region | EXP | 4345202 |8300609 | |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005737 | cytoplasm | TAS | 9622609 | |
| GO:0005886 | plasma membrane | EXP | 8300609 | |
| GO:0034364 | high-density lipoprotein particle | IDA | 210174 | |
| GO:0034362 | low-density lipoprotein particle | IDA | 8245722 | |
| GO:0034363 | intermediate-density lipoprotein particle | IDA | 17336988 | |
| GO:0034361 | very-low-density lipoprotein particle | IDA | 8245722 | |
| GO:0042627 | chylomicron | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| A2M | CPAMD5 | DKFZp779B086 | FWP007 | S863-7 | alpha-2-macroglobulin | - | HPRD,BioGRID | 9831625 |
| ALB | DKFZp779N1935 | PRO0883 | PRO0903 | PRO1341 | albumin | - | HPRD | 8063017 |
| APP | AAA | ABETA | ABPP | AD1 | APPI | CTFgamma | CVAP | PN2 | amyloid beta (A4) precursor protein | - | HPRD | 10559559 |
| CNTF | HCNTF | ciliary neurotrophic factor | Apolipoprotein E binds to ciliary neurotrophic factor | BIND | 9236223 |
| CTSB | APPS | CPSB | cathepsin B | - | HPRD | 12057992 |
| LDLR | FH | FHC | low density lipoprotein receptor | - | HPRD | 12036962 |
| LRP1 | A2MR | APOER | APR | CD91 | FLJ16451 | IGFBP3R | LRP | MGC88725 | TGFBR5 | low density lipoprotein-related protein 1 (alpha-2-macroglobulin receptor) | - | HPRD | 11421580 |
| LRP2 | DBS | gp330 | low density lipoprotein-related protein 2 | - | HPRD | 11421580 |
| LRP2 | DBS | gp330 | low density lipoprotein-related protein 2 | Reconstituted Complex | BioGRID | 7768901 |
| LRP8 | APOER2 | HSZ75190 | MCI1 | low density lipoprotein receptor-related protein 8, apolipoprotein e receptor | - | HPRD,BioGRID | 12167620 |12950167 |
| MAP2 | DKFZp686I2148 | MAP2A | MAP2B | MAP2C | microtubule-associated protein 2 | - | HPRD | 8624078 |
| MAPT | DDPAC | FLJ31424 | FTDP-17 | MAPTL | MGC138549 | MSTD | MTBT1 | MTBT2 | PPND | TAU | microtubule-associated protein tau | Reconstituted Complex | BioGRID | 8620924 |
| MAPT | DDPAC | FLJ31424 | FTDP-17 | MAPTL | MGC138549 | MSTD | MTBT1 | MTBT2 | PPND | TAU | microtubule-associated protein tau | - | HPRD | 7566652 |9147408 |
| NEFM | NEF3 | NF-M | NFM | neurofilament, medium polypeptide | Reconstituted Complex | BioGRID | 8620924 |
| PLTP | HDLCQ9 | phospholipid transfer protein | - | HPRD,BioGRID | 12810820 |
| SCARB1 | CD36L1 | CLA-1 | CLA1 | MGC138242 | SR-BI | SRB1 | scavenger receptor class B, member 1 | - | HPRD | 12138091 |
| SDC2 | HSPG | HSPG1 | SYND2 | syndecan 2 | - | HPRD | 9869645 |
| VLDLR | CHRMQ1 | FLJ35024 | VLDLRCH | very low density lipoprotein receptor | - | HPRD | 12167620 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ALZHEIMERS DISEASE | 169 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME HDL MEDIATED LIPID TRANSPORT | 15 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | 46 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME LIPOPROTEIN METABOLISM | 28 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | 16 | 15 | All SZGR 2.0 genes in this pathway |
| NAKAMURA CANCER MICROENVIRONMENT UP | 26 | 14 | All SZGR 2.0 genes in this pathway |
| PICCALUGA ANGIOIMMUNOBLASTIC LYMPHOMA UP | 205 | 140 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| BERTUCCI MEDULLARY VS DUCTAL BREAST CANCER UP | 206 | 111 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE UP | 351 | 230 | All SZGR 2.0 genes in this pathway |
| FOURNIER ACINAR DEVELOPMENT LATE UP | 11 | 6 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA MYELOID DN | 38 | 25 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 10D DN | 142 | 90 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 16D DN | 143 | 83 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC UP | 185 | 126 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION MONOCYTE UP | 204 | 140 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN | 459 | 276 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML QUIESCENT VS NORMAL QUIESCENT UP | 87 | 45 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML QUIESCENT VS NORMAL DIVIDING UP | 57 | 33 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 CHRONIC LOF DN | 118 | 78 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL DN | 186 | 107 | All SZGR 2.0 genes in this pathway |
| DARWICHE SKIN TUMOR PROMOTER DN | 185 | 115 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK LOW DN | 165 | 107 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK HIGH DN | 180 | 110 | All SZGR 2.0 genes in this pathway |
| DARWICHE SQUAMOUS CELL CARCINOMA DN | 181 | 107 | All SZGR 2.0 genes in this pathway |
| ROVERSI GLIOMA COPY NUMBER UP | 100 | 75 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER DN | 514 | 319 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 2 DN | 51 | 42 | All SZGR 2.0 genes in this pathway |
| MORI MATURE B LYMPHOCYTE UP | 90 | 62 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 1 UP | 140 | 85 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO DN | 200 | 112 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| ROSS AML OF FAB M7 TYPE | 68 | 44 | All SZGR 2.0 genes in this pathway |
| YAO HOXA10 TARGETS VIA PROGESTERONE UP | 79 | 58 | All SZGR 2.0 genes in this pathway |
| HSIAO LIVER SPECIFIC GENES | 244 | 154 | All SZGR 2.0 genes in this pathway |
| NADLER OBESITY DN | 48 | 34 | All SZGR 2.0 genes in this pathway |
| LENAOUR DENDRITIC CELL MATURATION UP | 114 | 84 | All SZGR 2.0 genes in this pathway |
| ABRAHAM ALPC VS MULTIPLE MYELOMA UP | 26 | 22 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 1 UP | 182 | 119 | All SZGR 2.0 genes in this pathway |
| APPEL IMATINIB RESPONSE | 33 | 22 | All SZGR 2.0 genes in this pathway |
| IGLESIAS E2F TARGETS UP | 151 | 103 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA DN | 41 | 27 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR UP | 240 | 152 | All SZGR 2.0 genes in this pathway |
| ABE VEGFA TARGETS 2HR | 34 | 21 | All SZGR 2.0 genes in this pathway |
| JIANG AGING HYPOTHALAMUS DN | 40 | 31 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 8HR UP | 105 | 73 | All SZGR 2.0 genes in this pathway |
| KRASNOSELSKAYA ILF3 TARGETS UP | 38 | 24 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR UP | 156 | 101 | All SZGR 2.0 genes in this pathway |
| YAMAZAKI TCEB3 TARGETS DN | 215 | 132 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK UP | 244 | 151 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 8 HR DN | 53 | 40 | All SZGR 2.0 genes in this pathway |
| MCLACHLAN DENTAL CARIES DN | 245 | 144 | All SZGR 2.0 genes in this pathway |
| LEE AGING CEREBELLUM UP | 84 | 58 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP | 487 | 303 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL DN | 128 | 93 | All SZGR 2.0 genes in this pathway |
| ABE VEGFA TARGETS 30MIN | 29 | 21 | All SZGR 2.0 genes in this pathway |
| JIANG AGING CEREBRAL CORTEX DN | 54 | 43 | All SZGR 2.0 genes in this pathway |
| MCLACHLAN DENTAL CARIES UP | 253 | 147 | All SZGR 2.0 genes in this pathway |
| GERHOLD ADIPOGENESIS UP | 49 | 40 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 6 | 189 | 112 | All SZGR 2.0 genes in this pathway |
| LEIN ASTROCYTE MARKERS | 42 | 35 | All SZGR 2.0 genes in this pathway |
| JEPSEN SMRT TARGETS | 33 | 23 | All SZGR 2.0 genes in this pathway |
| HELLER SILENCED BY METHYLATION UP | 282 | 183 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| WALLACE PROSTATE CANCER RACE UP | 299 | 167 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL UP | 260 | 174 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH DN | 31 | 25 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL CARCINOMA VS ADENOMA DN | 24 | 18 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C DN | 59 | 39 | All SZGR 2.0 genes in this pathway |
| HELLEBREKERS SILENCED DURING TUMOR ANGIOGENESIS | 80 | 56 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| RUIZ TNC TARGETS UP | 153 | 107 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP | 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| HOFFMANN IMMATURE TO MATURE B LYMPHOCYTE UP | 43 | 34 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR UP | 293 | 203 | All SZGR 2.0 genes in this pathway |
| MELLMAN TUT1 TARGETS DN | 47 | 29 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT DN | 222 | 141 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| GERHOLD RESPONSE TO TZD DN | 13 | 11 | All SZGR 2.0 genes in this pathway |
| ONGUSAHA BRCA1 TARGETS UP | 13 | 11 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 2HR UP | 39 | 30 | All SZGR 2.0 genes in this pathway |
| VANDESLUIS COMMD1 TARGETS GROUP 4 UP | 19 | 12 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION TOP20 UP | 20 | 14 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION UP | 271 | 165 | All SZGR 2.0 genes in this pathway |
| KATSANOU ELAVL1 TARGETS UP | 169 | 105 | All SZGR 2.0 genes in this pathway |
| SERVITJA ISLET HNF1A TARGETS UP | 163 | 111 | All SZGR 2.0 genes in this pathway |
| PECE MAMMARY STEM CELL UP | 146 | 75 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY LUMINAL MATURE DN | 99 | 74 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |