Gene Page: ITGB1
Summary ?
| GeneID | 3688 |
| Symbol | ITGB1 |
| Synonyms | CD29|FNRB|GPIIA|MDF2|MSK12|VLA-BETA|VLAB |
| Description | integrin subunit beta 1 |
| Reference | MIM:135630|HGNC:HGNC:6153|Ensembl:ENSG00000150093|HPRD:00628|Vega:OTTHUMG00000017928 |
| Gene type | protein-coding |
| Map location | 10p11.2 |
| Pascal p-value | 0.06 |
| Sherlock p-value | 0.624 |
| Fetal beta | 0.98 |
| eGene | Caudate basal ganglia |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0109 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
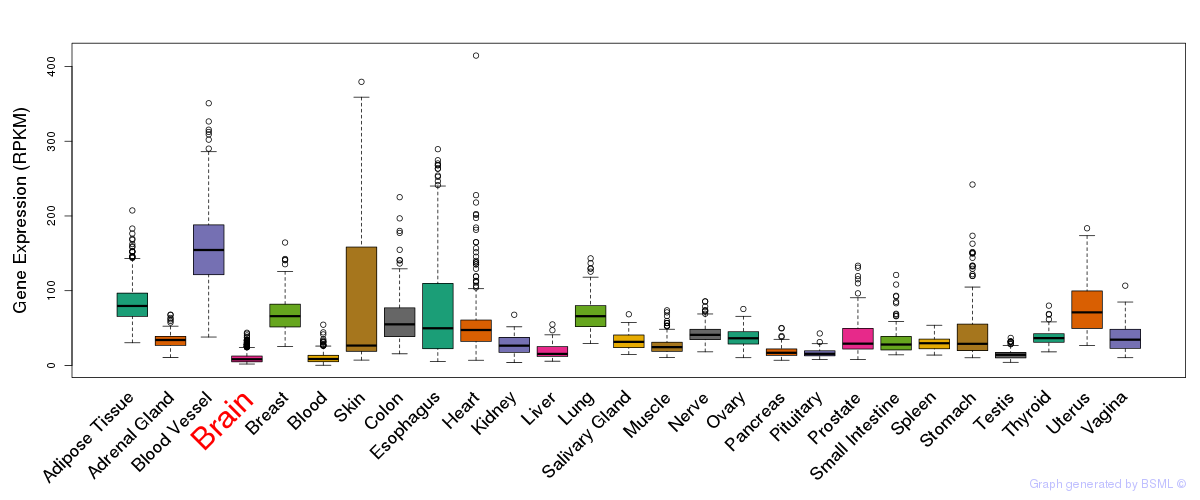
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004872 | receptor activity | IEA | - | |
| GO:0005178 | integrin binding | IEA | - | |
| GO:0005488 | binding | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0042802 | identical protein binding | IPI | 14681217 | |
| GO:0046982 | protein heterodimerization activity | NAS | 9552005 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000082 | G1/S transition of mitotic cell cycle | IEA | - | |
| GO:0001701 | in utero embryonic development | IEA | - | |
| GO:0007160 | cell-matrix adhesion | IEA | - | |
| GO:0007155 | cell adhesion | IEA | - | |
| GO:0007156 | homophilic cell adhesion | TAS | 10201960 | |
| GO:0007229 | integrin-mediated signaling pathway | IEA | - | |
| GO:0006968 | cellular defense response | TAS | 10201960 | |
| GO:0008284 | positive regulation of cell proliferation | IEA | - | |
| GO:0010002 | cardioblast differentiation | IEA | - | |
| GO:0007159 | leukocyte adhesion | IDA | 1715889 | |
| GO:0016477 | cell migration | TAS | 11919189 | |
| GO:0042685 | cardioblast cell fate specification | IEA | - | |
| GO:0030183 | B cell differentiation | IC | 1715889 | |
| GO:0044419 | interspecies interaction between organisms | IEA | - | |
| GO:0045596 | negative regulation of cell differentiation | IEA | - | |
| GO:0045214 | sarcomere organization | IEA | - | |
| GO:0051726 | regulation of cell cycle | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0031594 | neuromuscular junction | IDA | neuron, axon, Synap, Neurotransmitter (GO term level: 3) | 9415431 |
| GO:0031594 | neuromuscular junction | IEA | neuron, axon, Synap, Neurotransmitter (GO term level: 3) | - |
| GO:0019717 | synaptosome | IEA | Synap, Brain (GO term level: 7) | - |
| GO:0001726 | ruffle | TAS | 11919189 | |
| GO:0005783 | endoplasmic reticulum | IDA | 11256614 | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0009986 | cell surface | IDA | 1715889 | |
| GO:0008305 | integrin complex | IEA | - | |
| GO:0008305 | integrin complex | NAS | 9552005 | |
| GO:0042383 | sarcolemma | IDA | 9415431 | |
| GO:0042383 | sarcolemma | IEA | - | |
| GO:0042470 | melanosome | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTN1 | FLJ40884 | actinin, alpha 1 | Reconstituted Complex | BioGRID | 2116421 |
| ACTN4 | ACTININ-4 | DKFZp686K23158 | FSGS | FSGS1 | actinin, alpha 4 | - | HPRD | 2116421 |
| ARHGAP5 | GFI2 | RhoGAP5 | p190-B | Rho GTPase activating protein 5 | - | HPRD | 8537347 |
| CANX | CNX | FLJ26570 | IP90 | P90 | calnexin | - | HPRD,BioGRID | 8163531 |
| CD151 | GP27 | MER2 | PETA-3 | RAPH | SFA1 | TSPAN24 | CD151 molecule (Raph blood group) | Affinity Capture-Western | BioGRID | 12175627 |
| CD36 | CHDS7 | FAT | GP3B | GP4 | GPIV | PASIV | SCARB3 | CD36 molecule (thrombospondin receptor) | Affinity Capture-Western | BioGRID | 11238109 |
| CD46 | MCP | MGC26544 | MIC10 | TLX | TRA2.10 | CD46 molecule, complement regulatory protein | - | HPRD,BioGRID | 10741407 |
| CD47 | IAP | MER6 | OA3 | CD47 molecule | Affinity Capture-Western | BioGRID | 10397731 |
| CD63 | LAMP-3 | ME491 | MLA1 | OMA81H | TSPAN30 | CD63 molecule | Affinity Capture-Western | BioGRID | 12175627 |
| CD81 | S5.7 | TAPA1 | TSPAN28 | CD81 molecule | - | HPRD,BioGRID | 10229664 |
| CD82 | 4F9 | C33 | GR15 | IA4 | KAI1 | R2 | SAR2 | ST6 | TSPAN27 | CD82 molecule | - | HPRD | 8757325 |
| CD9 | 5H9 | BA2 | BTCC-1 | DRAP-27 | GIG2 | MIC3 | MRP-1 | P24 | TSPAN29 | CD9 molecule | Affinity Capture-Western | BioGRID | 8630057 |12175627 |
| COL1A1 | OI4 | collagen, type I, alpha 1 | Co-purification | BioGRID | 2156854 |
| FBXO2 | FBG1 | FBX2 | Fbs1 | NFB42 | F-box protein 2 | - | HPRD,BioGRID | 12140560 |
| FHL2 | AAG11 | DRAL | SLIM3 | four and a half LIM domains 2 | Affinity Capture-Western Two-hybrid | BioGRID | 10906324 |
| FLNA | ABP-280 | ABPX | DKFZp434P031 | FLN | FLN1 | FMD | MNS | NHBP | OPD | OPD1 | OPD2 | filamin A, alpha (actin binding protein 280) | - | HPRD,BioGRID | 9722563 |11807098 |
| FLNB | ABP-278 | AOI | DKFZp686A1668 | DKFZp686O033 | FH1 | FLN1L | LRS1 | SCT | TABP | TAP | filamin B, beta (actin binding protein 278) | - | HPRD,BioGRID | 11807098 |
| FLT4 | FLT41 | LMPH1A | PCL | VEGFR3 | fms-related tyrosine kinase 4 | - | HPRD,BioGRID | 11553610 |
| GNB2L1 | Gnb2-rs1 | H12.3 | HLC-7 | PIG21 | RACK1 | guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 | beta-1 integrin interacts with RACK1. | BIND | 11884618 |
| GNB2L1 | Gnb2-rs1 | H12.3 | HLC-7 | PIG21 | RACK1 | guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 | - | HPRD,BioGRID | 9442085 |
| GNB2L1 | Gnb2-rs1 | H12.3 | HLC-7 | PIG21 | RACK1 | guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 | An unspecified isoform of integrin-beta-1 interacts with RACK1. | BIND | 15611085 |
| HSPG2 | PLC | PRCAN | SJA | SJS | SJS1 | heparan sulfate proteoglycan 2 | - | HPRD | 9431988 |
| ICAM4 | CD242 | LW | intercellular adhesion molecule 4 (Landsteiner-Wiener blood group) | - | HPRD | 11435317 |
| IGF1R | CD221 | IGFIR | JTK13 | MGC142170 | MGC142172 | MGC18216 | insulin-like growth factor 1 receptor | beta-1 integrin interacts with IGF-IR. | BIND | 11884618 |
| ILK | DKFZp686F1765 | P59 | integrin-linked kinase | Integrin-beta-1 interacts with ILK. | BIND | 8538749 |
| ILK | DKFZp686F1765 | P59 | integrin-linked kinase | - | HPRD,BioGRID | 8538749 |9736715 |
| ITGA1 | CD49a | VLA1 | integrin, alpha 1 | - | HPRD | 8428973 |
| ITGA1 | CD49a | VLA1 | integrin, alpha 1 | VLA1-alpha1 interacts with VLA1-beta. | BIND | 1690718 |1693624 |3546305 |
| ITGA10 | PRO827 | integrin, alpha 10 | - | HPRD,BioGRID | 9685391 |
| ITGA11 | HsT18964 | integrin, alpha 11 | Integrin-alpha11 interacts with Integrin-beta1. | BIND | 10464311 |
| ITGA11 | HsT18964 | integrin, alpha 11 | - | HPRD | 10486209 |
| ITGA2 | BR | CD49B | GPIa | VLA-2 | VLAA2 | integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) | VLA2-alpha2 interacts with VLA2-beta. | BIND | 1690718 |1693624 |3546305 |
| ITGA3 | CD49C | FLJ34631 | FLJ34704 | GAP-B3 | GAPB3 | MSK18 | VCA-2 | VL3A | VLA3a | integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) | VLA3-alpha3 interacts with VLA3-beta. | BIND | 1690718 |1693624 |3546305 |
| ITGA3 | CD49C | FLJ34631 | FLJ34704 | GAP-B3 | GAPB3 | MSK18 | VCA-2 | VL3A | VLA3a | integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) | VLA3-alpha3 interacts with VLA3-beta | BIND | 3546305 |
| ITGA3 | CD49C | FLJ34631 | FLJ34704 | GAP-B3 | GAPB3 | MSK18 | VCA-2 | VL3A | VLA3a | integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) | - | HPRD,BioGRID | 15254262 |
| ITGA4 | CD49D | IA4 | MGC90518 | integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) | VLA4-alpha4 interacts with VLA4-beta. | BIND | 1693624 |3546305 |
| ITGA5 | CD49e | FNRA | VLA5A | integrin, alpha 5 (fibronectin receptor, alpha polypeptide) | VLA5-alpha5 interacts with VLA5-beta. | BIND | 1690718 |1693624 |3546305 |
| ITGA6 | CD49f | DKFZp686J01244 | FLJ18737 | ITGA6B | VLA-6 | integrin, alpha 6 | Affinity Capture-Western | BioGRID | 9660880 |
| ITGA6 | CD49f | DKFZp686J01244 | FLJ18737 | ITGA6B | VLA-6 | integrin, alpha 6 | - | HPRD | 9425259 |
| ITGA6 | CD49f | DKFZp686J01244 | FLJ18737 | ITGA6B | VLA-6 | integrin, alpha 6 | VLA6-alpha6 interacts with VLA-beta1. | BIND | 1690718 |1693624 |2138612 |2649503 |2967289 |
| ITGA8 | - | integrin, alpha 8 | - | HPRD,BioGRID | 7768999 |
| ITGA8 | - | integrin, alpha 8 | Integrin-alpha8 interacts with Integrin-beta1. | BIND | 7559467 |
| ITGA9 | ALPHA-RLC | ITGA4L | RLC | integrin, alpha 9 | Integrin-alpha9 interacts with Integrin-beta1. | BIND | 8798654 |
| ITGA9 | ALPHA-RLC | ITGA4L | RLC | integrin, alpha 9 | - | HPRD,BioGRID | 8245132|11462216 |
| ITGAV | CD51 | DKFZp686A08142 | MSK8 | VNRA | integrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51) | Integrin-alphaV interacts with Integrin-beta1 | BIND | 1690718 |2138612 |
| ITGB1BP1 | DKFZp686K08158 | ICAP-1A | ICAP-1B | ICAP-1alpha | ICAP1 | ICAP1A | ICAP1B | integrin beta 1 binding protein 1 | - | HPRD,BioGRID | 9281591 |11741908 |
| ITGB1BP2 | CHORDC3 | ITGB1BP | MELUSIN | MGC119214 | MSTP015 | integrin beta 1 binding protein (melusin) 2 | - | HPRD | 12514714 |
| ITGB1BP3 | MGC126624 | MIBP | NRK2 | integrin beta 1 binding protein 3 | - | HPRD | 10613898 |
| ITGB5 | FLJ26658 | integrin, beta 5 | - | HPRD | 11462216 |
| LAMA1 | LAMA | laminin, alpha 1 | - | HPRD,BioGRID | 9688542 |
| LGALS3BP | 90K | BTBD17B | MAC-2-BP | lectin, galactoside-binding, soluble, 3 binding protein | - | HPRD | 9501082 |
| LGALS8 | Gal-8 | PCTA-1 | PCTA1 | Po66-CBP | lectin, galactoside-binding, soluble, 8 | Affinity Capture-Western Reconstituted Complex | BioGRID | 10852818 |
| MAP4K4 | FLH21957 | FLJ10410 | FLJ20373 | FLJ90111 | HGK | KIAA0687 | NIK | mitogen-activated protein kinase kinase kinase kinase 4 | - | HPRD,BioGRID | 11967148 |
| MYO10 | FLJ10639 | FLJ21066 | FLJ22268 | FLJ43256 | KIAA0799 | MGC131988 | myosin X | Integrin-beta-1 interacts with Myo10. This interaction was modeled on a demonstrated interaction between human integrin-beta-1 and bovine Myo10. | BIND | 15156152 |
| NME1 | AWD | GAAD | NB | NBS | NDPK-A | NDPKA | NM23 | NM23-H1 | non-metastatic cells 1, protein (NM23A) expressed in | - | HPRD,BioGRID | 11919189 |
| PRKCA | AAG6 | MGC129900 | MGC129901 | PKC-alpha | PKCA | PRKACA | protein kinase C, alpha | - | HPRD,BioGRID | 12138200 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | - | HPRD | 8649427 |
| PXN | FLJ16691 | paxillin | - | HPRD,BioGRID | 7657702|10804218 |
| RAB8B | FLJ38125 | RAB8B, member RAS oncogene family | Affinity Capture-Western | BioGRID | 12639940 |
| SLC3A2 | 4F2 | 4F2HC | 4T2HC | CD98 | CD98HC | MDU1 | NACAE | solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 | - | HPRD,BioGRID | 12181350 |
| TGOLN2 | MGC14722 | TGN38 | TGN46 | TGN48 | TGN51 | TTGN2 | trans-golgi network protein 2 | - | HPRD,BioGRID | 11208159 |
| THBS1 | THBS | TSP | TSP1 | thrombospondin 1 | - | HPRD | 11980922 |
| TLN1 | ILWEQ | KIAA1027 | TLN | talin 1 | - | HPRD,BioGRID | 11279249 |
| TSPAN4 | NAG-2 | NAG2 | TETRASPAN | TM4SF7 | TSPAN-4 | tetraspanin 4 | - | HPRD,BioGRID | 9360996 |10734060 |
| VCAM1 | CD106 | DKFZp779G2333 | INCAM-100 | MGC99561 | vascular cell adhesion molecule 1 | - | HPRD | 12097820 |
| VCAN | CSPG2 | DKFZp686K06110 | ERVR | PG-M | WGN | WGN1 | versican | - | HPRD,BioGRID | 11805102 |
| YWHAB | GW128 | HS1 | KCIP-1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | - | HPRD,BioGRID | 11313964 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG AXON GUIDANCE | 129 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG FOCAL ADHESION | 201 | 138 | All SZGR 2.0 genes in this pathway |
| KEGG ECM RECEPTOR INTERACTION | 84 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG CELL ADHESION MOLECULES CAMS | 134 | 93 | All SZGR 2.0 genes in this pathway |
| KEGG LEUKOCYTE TRANSENDOTHELIAL MIGRATION | 118 | 78 | All SZGR 2.0 genes in this pathway |
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| KEGG PATHOGENIC ESCHERICHIA COLI INFECTION | 59 | 36 | All SZGR 2.0 genes in this pathway |
| KEGG LEISHMANIA INFECTION | 72 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG SMALL CELL LUNG CANCER | 84 | 67 | All SZGR 2.0 genes in this pathway |
| KEGG HYPERTROPHIC CARDIOMYOPATHY HCM | 85 | 65 | All SZGR 2.0 genes in this pathway |
| KEGG ARRHYTHMOGENIC RIGHT VENTRICULAR CARDIOMYOPATHY ARVC | 76 | 59 | All SZGR 2.0 genes in this pathway |
| KEGG DILATED CARDIOMYOPATHY | 92 | 68 | All SZGR 2.0 genes in this pathway |
| BIOCARTA LYM PATHWAY | 11 | 6 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AGR PATHWAY | 36 | 31 | All SZGR 2.0 genes in this pathway |
| BIOCARTA SPPA PATHWAY | 22 | 18 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BCELLSURVIVAL PATHWAY | 16 | 10 | All SZGR 2.0 genes in this pathway |
| BIOCARTA LAIR PATHWAY | 17 | 12 | All SZGR 2.0 genes in this pathway |
| BIOCARTA EPHA4 PATHWAY | 10 | 8 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ECM PATHWAY | 24 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ERK PATHWAY | 28 | 22 | All SZGR 2.0 genes in this pathway |
| BIOCARTA INTEGRIN PATHWAY | 38 | 29 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MCALPAIN PATHWAY | 25 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MONOCYTE PATHWAY | 11 | 5 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PTEN PATHWAY | 18 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NKCELLS PATHWAY | 20 | 13 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MET PATHWAY | 37 | 30 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TFF PATHWAY | 21 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA UCALPAIN PATHWAY | 18 | 11 | All SZGR 2.0 genes in this pathway |
| PID PRL SIGNALING EVENTS PATHWAY | 23 | 17 | All SZGR 2.0 genes in this pathway |
| PID RHOA PATHWAY | 45 | 33 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN1 PATHWAY | 66 | 44 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN CS PATHWAY | 26 | 16 | All SZGR 2.0 genes in this pathway |
| PID ARF6 TRAFFICKING PATHWAY | 49 | 34 | All SZGR 2.0 genes in this pathway |
| PID REELIN PATHWAY | 29 | 29 | All SZGR 2.0 genes in this pathway |
| PID TCPTP PATHWAY | 43 | 33 | All SZGR 2.0 genes in this pathway |
| PID ANGIOPOIETIN RECEPTOR PATHWAY | 50 | 41 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN A9B1 PATHWAY | 25 | 18 | All SZGR 2.0 genes in this pathway |
| PID CXCR4 PATHWAY | 102 | 78 | All SZGR 2.0 genes in this pathway |
| PID SYNDECAN 4 PATHWAY | 32 | 25 | All SZGR 2.0 genes in this pathway |
| PID UPA UPAR PATHWAY | 42 | 30 | All SZGR 2.0 genes in this pathway |
| PID A6B1 A6B4 INTEGRIN PATHWAY | 46 | 35 | All SZGR 2.0 genes in this pathway |
| PID SYNDECAN 2 PATHWAY | 33 | 27 | All SZGR 2.0 genes in this pathway |
| PID MYC REPRESS PATHWAY | 63 | 52 | All SZGR 2.0 genes in this pathway |
| PID LYMPH ANGIOGENESIS PATHWAY | 25 | 16 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN A4B1 PATHWAY | 33 | 24 | All SZGR 2.0 genes in this pathway |
| PID FAK PATHWAY | 59 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CELL COMMUNICATION | 120 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | 70 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | 91 | 65 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | 12 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME BASIGIN INTERACTIONS | 30 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRIN CELL SURFACE INTERACTIONS | 79 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME AXON GUIDANCE | 251 | 188 | All SZGR 2.0 genes in this pathway |
| REACTOME OTHER SEMAPHORIN INTERACTIONS | 15 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME SEMAPHORIN INTERACTIONS | 68 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | 14 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL JUNCTION ORGANIZATION | 78 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME L1CAM INTERACTIONS | 86 | 62 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNAL TRANSDUCTION BY L1 | 34 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| WATANABE RECTAL CANCER RADIOTHERAPY RESPONSIVE DN | 92 | 51 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 UP | 201 | 125 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL DN | 455 | 304 | All SZGR 2.0 genes in this pathway |
| AKL HTLV1 INFECTION DN | 68 | 41 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS UP | 457 | 269 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS UP | 238 | 144 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC2 TARGETS UP | 114 | 66 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN | 459 | 276 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED UP | 633 | 376 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| HUMMERICH SKIN CANCER PROGRESSION UP | 88 | 58 | All SZGR 2.0 genes in this pathway |
| SASAKI TARGETS OF TP73 AND TP63 | 11 | 7 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| LIU BREAST CANCER | 30 | 19 | All SZGR 2.0 genes in this pathway |
| GUENTHER GROWTH SPHERICAL VS ADHERENT DN | 26 | 19 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION UP | 69 | 55 | All SZGR 2.0 genes in this pathway |
| HE PTEN TARGETS UP | 16 | 15 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST DN | 309 | 206 | All SZGR 2.0 genes in this pathway |
| FERNANDEZ BOUND BY MYC | 182 | 116 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT OK VS DONOR UP | 151 | 100 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT REJECTED VS OK DN | 51 | 35 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| IGLESIAS E2F TARGETS UP | 151 | 103 | All SZGR 2.0 genes in this pathway |
| ZHOU TNF SIGNALING 4HR | 54 | 36 | All SZGR 2.0 genes in this pathway |
| RAMALHO STEMNESS UP | 206 | 118 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| YAMAZAKI TCEB3 TARGETS UP | 175 | 116 | All SZGR 2.0 genes in this pathway |
| ZHANG PROLIFERATING VS QUIESCENT | 51 | 41 | All SZGR 2.0 genes in this pathway |
| GERHOLD ADIPOGENESIS DN | 64 | 44 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN UP | 262 | 186 | All SZGR 2.0 genes in this pathway |
| HENDRICKS SMARCA4 TARGETS DN | 53 | 34 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C7 | 68 | 44 | All SZGR 2.0 genes in this pathway |
| DURCHDEWALD SKIN CARCINOGENESIS DN | 264 | 168 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER T4 | 94 | 69 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY 4NQO OR UV | 63 | 44 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE DN | 195 | 135 | All SZGR 2.0 genes in this pathway |
| AMUNDSON POOR SURVIVAL AFTER GAMMA RADIATION 2G | 171 | 96 | All SZGR 2.0 genes in this pathway |
| LABBE TGFB1 TARGETS UP | 102 | 64 | All SZGR 2.0 genes in this pathway |
| LABBE TARGETS OF TGFB1 AND WNT3A UP | 111 | 70 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA UP | 259 | 185 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE DN | 264 | 159 | All SZGR 2.0 genes in this pathway |
| GOLDRATH ANTIGEN RESPONSE | 346 | 192 | All SZGR 2.0 genes in this pathway |
| MELLMAN TUT1 TARGETS DN | 47 | 29 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| LIU VAV3 PROSTATE CARCINOGENESIS DN | 17 | 14 | All SZGR 2.0 genes in this pathway |
| KAPOSI LIVER CANCER MET UP | 18 | 11 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN | 253 | 192 | All SZGR 2.0 genes in this pathway |
| DANG MYC TARGETS DN | 31 | 25 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| GREGORY SYNTHETIC LETHAL WITH IMATINIB | 145 | 83 | All SZGR 2.0 genes in this pathway |
| KANG AR TARGETS DN | 19 | 12 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS DN | 315 | 215 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 7 | 403 | 240 | All SZGR 2.0 genes in this pathway |
| PEDERSEN TARGETS OF 611CTF ISOFORM OF ERBB2 | 76 | 45 | All SZGR 2.0 genes in this pathway |
| ABDELMOHSEN ELAVL4 TARGETS | 16 | 13 | All SZGR 2.0 genes in this pathway |
| PECE MAMMARY STEM CELL DN | 146 | 88 | All SZGR 2.0 genes in this pathway |
| KUMAR PATHOGEN LOAD BY MACROPHAGES | 275 | 155 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124/506 | 1095 | 1101 | m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA | ||||
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-134 | 52 | 58 | 1A | hsa-miR-134brain | UGUGACUGGUUGACCAGAGGG |
| miR-183 | 623 | 629 | m8 | hsa-miR-183 | UAUGGCACUGGUAGAAUUCACUG |
| miR-223 | 183 | 189 | 1A | hsa-miR-223 | UGUCAGUUUGUCAAAUACCCC |
| miR-29 | 830 | 836 | m8 | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
| miR-320 | 608 | 614 | 1A | hsa-miR-320 | AAAAGCUGGGUUGAGAGGGCGAA |
| miR-493-5p | 115 | 122 | 1A,m8 | hsa-miR-493-5p | UUGUACAUGGUAGGCUUUCAUU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.