Gene Page: MCM7
Summary ?
| GeneID | 4176 |
| Symbol | MCM7 |
| Synonyms | CDC47|MCM2|P1.1-MCM3|P1CDC47|P85MCM|PNAS146|PPP1R104 |
| Description | minichromosome maintenance complex component 7 |
| Reference | MIM:600592|HGNC:HGNC:6950|Ensembl:ENSG00000166508|HPRD:01154|Vega:OTTHUMG00000154671 |
| Gene type | protein-coding |
| Map location | 7q21.3-q22.1 |
| Pascal p-value | 0.461 |
| Sherlock p-value | 8.163E-4 |
| Fetal beta | 2.271 |
| DMG | 2 (# studies) |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 2 |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 2 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.013 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg26023204 | 7 | 99699378 | MCM7 | -0.02 | 0.87 | DMG:Nishioka_2013 | |
| cg13650156 | 7 | 99970502 | MCM7 | 2.22E-6 | 3.895 | DMG:vanEijk_2014 |
Section II. Transcriptome annotation
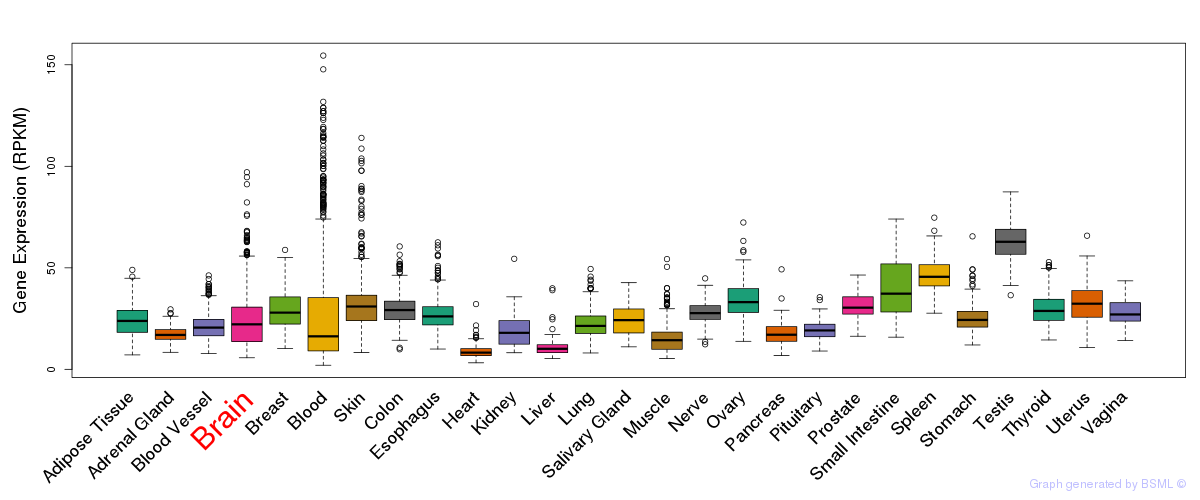
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CNPY2 | 0.86 | 0.81 |
| COPS6 | 0.86 | 0.84 |
| CCDC56 | 0.86 | 0.86 |
| MYL6 | 0.86 | 0.83 |
| CHMP2A | 0.86 | 0.83 |
| PSMA5 | 0.85 | 0.83 |
| NHP2 | 0.85 | 0.82 |
| SPAG7 | 0.85 | 0.85 |
| TOMM5 | 0.85 | 0.84 |
| TSTA3 | 0.84 | 0.81 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.26 | -0.62 | -0.56 |
| AF347015.8 | -0.59 | -0.51 |
| AF347015.2 | -0.58 | -0.51 |
| AF347015.15 | -0.58 | -0.53 |
| AF347015.18 | -0.58 | -0.51 |
| MT-CYB | -0.56 | -0.52 |
| MT-CO2 | -0.56 | -0.47 |
| AF347015.33 | -0.56 | -0.50 |
| AF347015.27 | -0.55 | -0.52 |
| MT-ATP8 | -0.55 | -0.52 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003677 | DNA binding | TAS | 15538388 | |
| GO:0005515 | protein binding | IPI | 10518787 |15538388 |16438930 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0017111 | nucleoside-triphosphatase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0006270 | DNA replication initiation | IEA | - | |
| GO:0006260 | DNA replication | EXP | 12791985 | |
| GO:0006260 | DNA replication | TAS | 8798650 | |
| GO:0007049 | cell cycle | IEA | - | |
| GO:0006974 | response to DNA damage stimulus | IMP | 15538388 | |
| GO:0042325 | regulation of phosphorylation | IMP | 15538388 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000785 | chromatin | TAS | 8798650 | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005654 | nucleoplasm | EXP | 10436018 |10846177 |11095689 |11931757 |12045100 |15226314 |15707391 | |
| GO:0005737 | cytoplasm | IDA | 18029348 | |
| GO:0042555 | MCM complex | IMP | 15538388 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ATR | FRP1 | MEC1 | SCKL | SCKL1 | ataxia telangiectasia and Rad3 related | Affinity Capture-Western | BioGRID | 15210935 |
| ATRIP | DKFZp762J2115 | FLJ12343 | MGC20625 | MGC21482 | MGC26740 | ATR interacting protein | Affinity Capture-Western | BioGRID | 15210935 |
| CCNH | CAK | p34 | p37 | cyclin H | - | HPRD,BioGRID | 11056214 |
| CDC45L | CDC45 | CDC45L2 | PORC-PI-1 | CDC45 cell division cycle 45-like (S. cerevisiae) | - | HPRD,BioGRID | 10518787 |12614612 |
| CDC6 | CDC18L | HsCDC18 | HsCDC6 | cell division cycle 6 homolog (S. cerevisiae) | - | HPRD,BioGRID | 10464337 |12614612 |
| CDC7 | CDC7L1 | HsCDC7 | Hsk1 | MGC117361 | MGC126237 | MGC126238 | huCDC7 | cell division cycle 7 homolog (S. cerevisiae) | - | HPRD,BioGRID | 12614612 |
| CDK7 | CAK1 | CDKN7 | MO15 | STK1 | p39MO15 | cyclin-dependent kinase 7 | - | HPRD,BioGRID | 11056214 |
| CNOT8 | CAF1 | CALIF | POP2 | hCAF1 | CCR4-NOT transcription complex, subunit 8 | Affinity Capture-MS | BioGRID | 17353931 |
| CRLS1 | C20orf155 | CLS1 | GCD10 | dJ967N21.6 | cardiolipin synthase 1 | - | HPRD | 12771218 |
| DBF4 | ASK | CHIF | DBF4A | ZDBF1 | DBF4 homolog (S. cerevisiae) | - | HPRD,BioGRID | 12614612 |
| FHL2 | AAG11 | DRAL | SLIM3 | four and a half LIM domains 2 | - | HPRD,BioGRID | 10649446 |
| GIPC1 | C19orf3 | GIPC | GLUT1CBP | Hs.6454 | IIP-1 | MGC15889 | MGC3774 | NIP | RGS19IP1 | SEMCAP | SYNECTIIN | SYNECTIN | TIP-2 | GIPC PDZ domain containing family, member 1 | Affinity Capture-MS | BioGRID | 17353931 |
| GSTK1 | GST13 | glutathione S-transferase kappa 1 | Affinity Capture-MS | BioGRID | 17353931 |
| HIST3H3 | H3.4 | H3/g | H3FT | H3t | MGC126886 | MGC126888 | histone cluster 3, H3 | - | HPRD,BioGRID | 8798650 |
| IKBKG | AMCBX1 | FIP-3 | FIP3 | Fip3p | IKK-gamma | IP | IP1 | IP2 | IPD2 | NEMO | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma | - | HPRD | 14743216 |
| MBIP | - | MAP3K12 binding inhibitory protein 1 | Two-hybrid | BioGRID | 16189514 |
| MCM10 | CNA43 | DNA43 | MGC126776 | PRO2249 | minichromosome maintenance complex component 10 | - | HPRD,BioGRID | 11095689 |15232106 |
| MCM2 | BM28 | CCNL1 | CDCL1 | D3S3194 | KIAA0030 | MGC10606 | MITOTIN | cdc19 | minichromosome maintenance complex component 2 | Affinity Capture-Western Co-purification in vivo Reconstituted Complex Two-hybrid | BioGRID | 8798650 |9099751 |10567526 |10748114 |12207017 |12614612 |12694531 |
| MCM2 | BM28 | CCNL1 | CDCL1 | D3S3194 | KIAA0030 | MGC10606 | MITOTIN | cdc19 | minichromosome maintenance complex component 2 | - | HPRD | 8798650 |10567526 |10748114 |12207017 |
| MCM2 | BM28 | CCNL1 | CDCL1 | D3S3194 | KIAA0030 | MGC10606 | MITOTIN | cdc19 | minichromosome maintenance complex component 2 | - | HPRD | 8798650 |10567526 |10748114 |12207017|15232106 |
| MCM3 | HCC5 | MGC1157 | P1-MCM3 | P1.h | RLFB | minichromosome maintenance complex component 3 | MCM3 interacts with MCM7. | BIND | 15236977 |
| MCM3 | HCC5 | MGC1157 | P1-MCM3 | P1.h | RLFB | minichromosome maintenance complex component 3 | - | HPRD | 7601140 |8631321 |10464337 |
| MCM3 | HCC5 | MGC1157 | P1-MCM3 | P1.h | RLFB | minichromosome maintenance complex component 3 | Mcm3 interacts with Mcm7. | BIND | 15236977 |
| MCM3 | HCC5 | MGC1157 | P1-MCM3 | P1.h | RLFB | minichromosome maintenance complex component 3 | Affinity Capture-Western Co-fractionation Two-hybrid | BioGRID | 9099751 |10464337 |11779870 |12614612 |
| MCM4 | CDC21 | CDC54 | MGC33310 | P1-CDC21 | hCdc21 | minichromosome maintenance complex component 4 | Affinity Capture-Western in vivo Reconstituted Complex | BioGRID | 8798650 |9099751 |10748114 |12207017 |12694531 |
| MCM4 | CDC21 | CDC54 | MGC33310 | P1-CDC21 | hCdc21 | minichromosome maintenance complex component 4 | Mcm4 interacts with Mcm7. | BIND | 15236977 |
| MCM4 | CDC21 | CDC54 | MGC33310 | P1-CDC21 | hCdc21 | minichromosome maintenance complex component 4 | - | HPRD | 7601140 |8631321 |8798650 |10567526 |10748114 |12207017 |
| MCM4 | CDC21 | CDC54 | MGC33310 | P1-CDC21 | hCdc21 | minichromosome maintenance complex component 4 | MCM4 interacts with MCM7. | BIND | 15236977 |
| MCM5 | CDC46 | MGC5315 | P1-CDC46 | minichromosome maintenance complex component 5 | Affinity Capture-Western Two-hybrid | BioGRID | 9099751 |11248027 |12614612 |
| MCM5 | CDC46 | MGC5315 | P1-CDC46 | minichromosome maintenance complex component 5 | - | HPRD | 9099751 |12614612 |
| MCM6 | MCG40308 | Mis5 | P105MCM | minichromosome maintenance complex component 6 | - | HPRD | 8798650 |10567526 |10748114 |12207017 |
| MCM6 | MCG40308 | Mis5 | P105MCM | minichromosome maintenance complex component 6 | Affinity Capture-Western in vivo Reconstituted Complex Two-hybrid | BioGRID | 8798650 |9099751 |10748114 |12207017 |12614612 |12694531 |
| MCM6 | MCG40308 | Mis5 | P105MCM | minichromosome maintenance complex component 6 | - | HPRD | 8798650 |10567526 |10748114 |12207017|12694531 |
| MCM7 | CDABP0042 | CDC47 | MCM2 | P1.1-MCM3 | P1CDC47 | P85MCM | PNAS-146 | minichromosome maintenance complex component 7 | Mcm7 interacts with itself. | BIND | 15236977 |
| MCM7 | CDABP0042 | CDC47 | MCM2 | P1.1-MCM3 | P1CDC47 | P85MCM | PNAS-146 | minichromosome maintenance complex component 7 | Reconstituted Complex Two-hybrid | BioGRID | 12614612 |12694531 |
| MCM8 | C20orf154 | MGC119522 | MGC119523 | MGC12866 | MGC4816 | REC | dJ967N21.5 | minichromosome maintenance complex component 8 | - | HPRD,BioGRID | 12771218 |
| MNAT1 | MAT1 | RNF66 | TFB3 | menage a trois homolog 1, cyclin H assembly factor (Xenopus laevis) | - | HPRD,BioGRID | 11056214 |
| MYC | bHLHe39 | c-Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | Affinity Capture-MS | BioGRID | 17353931 |
| NEK6 | SID6-1512 | NIMA (never in mitosis gene a)-related kinase 6 | Affinity Capture-MS | BioGRID | 17353931 |
| NFKBIA | IKBA | MAD-3 | NFKBI | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | - | HPRD | 14743216 |
| ORC1L | HSORC1 | ORC1 | PARC1 | origin recognition complex, subunit 1-like (yeast) | - | HPRD,BioGRID | 12614612 |
| ORC2L | ORC2 | origin recognition complex, subunit 2-like (yeast) | - | HPRD,BioGRID | 12614612 |
| ORC3L | LAT | LATHEO | ORC3 | origin recognition complex, subunit 3-like (yeast) | - | HPRD,BioGRID | 12614612 |
| ORC5L | ORC5 | ORC5P | ORC5T | origin recognition complex, subunit 5-like (yeast) | - | HPRD,BioGRID | 12614612 |
| PELO | CGI-17 | PRO1770 | pelota homolog (Drosophila) | Affinity Capture-MS | BioGRID | 17353931 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | - | HPRD,BioGRID | 9566894 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | MCM7 interacts with Rb. | BIND | 9566894 |
| RBL1 | CP107 | MGC40006 | PRB1 | p107 | retinoblastoma-like 1 (p107) | - | HPRD,BioGRID | 9566894 |
| RBL1 | CP107 | MGC40006 | PRB1 | p107 | retinoblastoma-like 1 (p107) | MCM7 interacts with p107. | BIND | 9566894 |
| RBL2 | FLJ26459 | P130 | Rb2 | retinoblastoma-like 2 (p130) | - | HPRD,BioGRID | 9566894 |
| RBL2 | FLJ26459 | P130 | Rb2 | retinoblastoma-like 2 (p130) | MCM7 interacts with p130. | BIND | 9566894 |
| RBM8A | BOV-1A | BOV-1B | BOV-1C | MDS014 | RBM8 | RBM8B | Y14 | ZNRP | ZRNP1 | RNA binding motif protein 8A | Two-hybrid | BioGRID | 16189514 |
| RPA1 | HSSB | REPA1 | RF-A | RP-A | RPA70 | replication protein A1, 70kDa | - | HPRD,BioGRID | 12614612 |
| TH1L | HSPC130 | NELF-C | NELF-D | TH1 | TH1-like (Drosophila) | Affinity Capture-MS | BioGRID | 17353931 |
| TRAF6 | MGC:3310 | RNF85 | TNF receptor-associated factor 6 | Affinity Capture-MS | BioGRID | 17353931 |
| TREX1 | AGS1 | AGS5 | CRV | DKFZp434J0310 | DRN3 | HERNS | three prime repair exonuclease 1 | - | HPRD | 15210935 |
| UBE3A | ANCR | AS | E6-AP | EPVE6AP | FLJ26981 | HPVE6A | ubiquitin protein ligase E3A | - | HPRD,BioGRID | 9852095 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG DNA REPLICATION | 36 | 21 | All SZGR 2.0 genes in this pathway |
| KEGG CELL CYCLE | 128 | 84 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MCM PATHWAY | 18 | 8 | All SZGR 2.0 genes in this pathway |
| PID ATR PATHWAY | 39 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | 31 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME ORC1 REMOVAL FROM CHROMATIN | 67 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE CHECKPOINTS | 124 | 70 | All SZGR 2.0 genes in this pathway |
| REACTOME M G1 TRANSITION | 81 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME G1 S TRANSITION | 112 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS OF DNA | 92 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC G1 G1 S PHASES | 137 | 79 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC M M G1 PHASES | 172 | 98 | All SZGR 2.0 genes in this pathway |
| REACTOME ASSEMBLY OF THE PRE REPLICATIVE COMPLEX | 65 | 36 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPLICATION | 192 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | 38 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME UNWINDING OF DNA | 11 | 6 | All SZGR 2.0 genes in this pathway |
| REACTOME G2 M CHECKPOINTS | 45 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME S PHASE | 109 | 66 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA STRAND ELONGATION | 30 | 18 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS DN | 310 | 188 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS UP | 290 | 177 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY UP | 430 | 232 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED UP | 633 | 376 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 CHRONIC LOF UP | 115 | 78 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF DN | 228 | 137 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| WANG ESOPHAGUS CANCER VS NORMAL UP | 121 | 72 | All SZGR 2.0 genes in this pathway |
| KAN RESPONSE TO ARSENIC TRIOXIDE | 123 | 80 | All SZGR 2.0 genes in this pathway |
| EGUCHI CELL CYCLE RB1 TARGETS | 23 | 13 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION | 77 | 51 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION HBZ | 41 | 27 | All SZGR 2.0 genes in this pathway |
| OXFORD RALA OR RALB TARGETS UP | 48 | 23 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN MELANOMA RELAPSE UP | 61 | 25 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| LOCKWOOD AMPLIFIED IN LUNG CANCER | 214 | 139 | All SZGR 2.0 genes in this pathway |
| SCIAN CELL CYCLE TARGETS OF TP53 AND TP73 DN | 22 | 14 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES 1 | 379 | 235 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| MORI PRE BI LYMPHOCYTE UP | 80 | 54 | All SZGR 2.0 genes in this pathway |
| MORI LARGE PRE BII LYMPHOCYTE UP | 86 | 49 | All SZGR 2.0 genes in this pathway |
| MORI IMMATURE B LYMPHOCYTE DN | 90 | 55 | All SZGR 2.0 genes in this pathway |
| MORI EMU MYC LYMPHOMA BY ONSET TIME UP | 110 | 69 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPLICATION GENES | 147 | 87 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER CIPROFIBRATE DN | 66 | 43 | All SZGR 2.0 genes in this pathway |
| ASTON MAJOR DEPRESSIVE DISORDER DN | 160 | 110 | All SZGR 2.0 genes in this pathway |
| LE EGR2 TARGETS UP | 108 | 75 | All SZGR 2.0 genes in this pathway |
| FERNANDEZ BOUND BY MYC | 182 | 116 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT REJECTED VS OK DN | 51 | 35 | All SZGR 2.0 genes in this pathway |
| SHIPP DLBCL VS FOLLICULAR LYMPHOMA UP | 45 | 30 | All SZGR 2.0 genes in this pathway |
| PEART HDAC PROLIFERATION CLUSTER DN | 76 | 57 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS UP | 126 | 84 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| LEI MYB TARGETS | 318 | 215 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS DN | 234 | 137 | All SZGR 2.0 genes in this pathway |
| VERNELL RETINOBLASTOMA PATHWAY UP | 70 | 47 | All SZGR 2.0 genes in this pathway |
| HESS TARGETS OF HOXA9 AND MEIS1 UP | 65 | 44 | All SZGR 2.0 genes in this pathway |
| IGLESIAS E2F TARGETS UP | 151 | 103 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER HEREDITARY VS SPORADIC | 50 | 32 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC DN | 228 | 146 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 3 | 101 | 64 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO CANTHARIDIN DN | 69 | 46 | All SZGR 2.0 genes in this pathway |
| ZHOU TNF SIGNALING 30MIN | 54 | 36 | All SZGR 2.0 genes in this pathway |
| SONG TARGETS OF IE86 CMV PROTEIN | 60 | 42 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER BRCA1 VS BRCA2 | 163 | 113 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 24HR UP | 148 | 96 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS PEAK AT 16HR | 40 | 24 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| GAVIN IL2 RESPONSIVE FOXP3 TARGETS UP | 19 | 12 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| SANSOM APC MYC TARGETS | 217 | 138 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS REQUIRE MYC | 210 | 123 | All SZGR 2.0 genes in this pathway |
| LEE METASTASIS AND ALTERNATIVE SPLICING UP | 74 | 51 | All SZGR 2.0 genes in this pathway |
| SARRIO EPITHELIAL MESENCHYMAL TRANSITION UP | 180 | 114 | All SZGR 2.0 genes in this pathway |
| FUJII YBX1 TARGETS DN | 202 | 132 | All SZGR 2.0 genes in this pathway |
| DAIRKEE CANCER PRONE RESPONSE BPA E2 | 118 | 65 | All SZGR 2.0 genes in this pathway |
| MOLENAAR TARGETS OF CCND1 AND CDK4 DN | 58 | 25 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| WHITEFORD PEDIATRIC CANCER MARKERS | 116 | 63 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| GRADE COLON AND RECTAL CANCER UP | 285 | 167 | All SZGR 2.0 genes in this pathway |
| RAY TUMORIGENESIS BY ERBB2 CDC25A UP | 104 | 57 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER SURVIVAL DN | 175 | 103 | All SZGR 2.0 genes in this pathway |
| HELLEBREKERS SILENCED DURING TUMOR ANGIOGENESIS | 80 | 56 | All SZGR 2.0 genes in this pathway |
| FRASOR RESPONSE TO SERM OR FULVESTRANT DN | 50 | 29 | All SZGR 2.0 genes in this pathway |
| RUIZ TNC TARGETS DN | 142 | 79 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE UP | 212 | 128 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR DN | 277 | 166 | All SZGR 2.0 genes in this pathway |
| CROONQUIST IL6 DEPRIVATION DN | 98 | 69 | All SZGR 2.0 genes in this pathway |
| CROONQUIST NRAS SIGNALING DN | 72 | 47 | All SZGR 2.0 genes in this pathway |
| CROONQUIST NRAS VS STROMAL STIMULATION DN | 99 | 65 | All SZGR 2.0 genes in this pathway |
| ISHIDA E2F TARGETS | 53 | 27 | All SZGR 2.0 genes in this pathway |
| KOBAYASHI EGFR SIGNALING 24HR DN | 251 | 151 | All SZGR 2.0 genes in this pathway |
| FOURNIER ACINAR DEVELOPMENT LATE 2 | 277 | 172 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| WONG EMBRYONIC STEM CELL CORE | 335 | 193 | All SZGR 2.0 genes in this pathway |
| WU APOPTOSIS BY CDKN1A VIA TP53 | 55 | 31 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS GROWING | 243 | 155 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 DN | 374 | 217 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 6HR UP | 229 | 149 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR UP | 324 | 193 | All SZGR 2.0 genes in this pathway |
| WIERENGA PML INTERACTOME | 42 | 23 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS VIA AKT1 UP | 281 | 183 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| ZHOU CELL CYCLE GENES IN IR RESPONSE 6HR | 85 | 49 | All SZGR 2.0 genes in this pathway |
| ZHOU CELL CYCLE GENES IN IR RESPONSE 24HR | 128 | 73 | All SZGR 2.0 genes in this pathway |