Gene Page: BDNF
Summary ?
| GeneID | 627 |
| Symbol | BDNF |
| Synonyms | ANON2|BULN2 |
| Description | brain-derived neurotrophic factor |
| Reference | MIM:113505|HGNC:HGNC:1033|Ensembl:ENSG00000176697|HPRD:00214|Vega:OTTHUMG00000178797 |
| Gene type | protein-coding |
| Map location | 11p13 |
| Pascal p-value | 0.002 |
| Sherlock p-value | 0.855 |
| Fetal beta | -1.443 |
| DMG | 1 (# studies) |
| eGene | Nucleus accumbens basal ganglia Putamen basal ganglia Myers' cis & trans |
| Support | NEUROTROPHIN SIGNALING Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 2 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizotypy,schizophrenias,schizotypal | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 10 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg10558494 | 11 | 27721280 | BDNF | 1.68E-8 | -0.011 | 6.13E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6474972 | 0 | BDNF | 627 | 0.19 | trans | |||
| rs12452652 | chr17 | 72235560 | BDNF | 627 | 0.09 | trans | ||
| rs11030023 | 11 | 27505351 | BDNF | ENSG00000176697.14 | 4.073E-6 | 0.03 | 238254 | gtex_brain_putamen_basal |
| rs10835187 | 11 | 27505677 | BDNF | ENSG00000176697.14 | 1.53E-6 | 0.03 | 237928 | gtex_brain_putamen_basal |
| rs112806008 | 11 | 27506369 | BDNF | ENSG00000176697.14 | 3.968E-6 | 0.03 | 237236 | gtex_brain_putamen_basal |
| rs10767644 | 11 | 27520671 | BDNF | ENSG00000176697.14 | 2.664E-6 | 0.03 | 222934 | gtex_brain_putamen_basal |
| rs11030036 | 11 | 27525124 | BDNF | ENSG00000176697.14 | 2.798E-6 | 0.03 | 218481 | gtex_brain_putamen_basal |
| rs12362567 | 11 | 27526645 | BDNF | ENSG00000176697.14 | 3.622E-6 | 0.03 | 216960 | gtex_brain_putamen_basal |
| rs3763965 | 11 | 27528987 | BDNF | ENSG00000176697.14 | 2.647E-6 | 0.03 | 214618 | gtex_brain_putamen_basal |
| rs67924215 | 11 | 27533660 | BDNF | ENSG00000176697.14 | 2.63E-6 | 0.03 | 209945 | gtex_brain_putamen_basal |
| rs10835189 | 11 | 27541995 | BDNF | ENSG00000176697.14 | 3.628E-6 | 0.03 | 201610 | gtex_brain_putamen_basal |
| rs11030042 | 11 | 27542328 | BDNF | ENSG00000176697.14 | 3.628E-6 | 0.03 | 201277 | gtex_brain_putamen_basal |
| rs4528277 | 11 | 27565446 | BDNF | ENSG00000176697.14 | 3.62E-6 | 0.03 | 178159 | gtex_brain_putamen_basal |
| rs11603350 | 11 | 27567246 | BDNF | ENSG00000176697.14 | 3.628E-6 | 0.03 | 176359 | gtex_brain_putamen_basal |
| rs901926 | 11 | 27570553 | BDNF | ENSG00000176697.14 | 3.628E-6 | 0.03 | 173052 | gtex_brain_putamen_basal |
| rs1304101 | 11 | 27571594 | BDNF | ENSG00000176697.14 | 3.628E-6 | 0.03 | 172011 | gtex_brain_putamen_basal |
| rs10835193 | 11 | 27572337 | BDNF | ENSG00000176697.14 | 3.974E-6 | 0.03 | 171268 | gtex_brain_putamen_basal |
| rs11030048 | 11 | 27582949 | BDNF | ENSG00000176697.14 | 3.628E-6 | 0.03 | 160656 | gtex_brain_putamen_basal |
| rs11030052 | 11 | 27591812 | BDNF | ENSG00000176697.14 | 3.63E-6 | 0.03 | 151793 | gtex_brain_putamen_basal |
| rs1387145 | 11 | 27600158 | BDNF | ENSG00000176697.14 | 3.635E-6 | 0.03 | 143447 | gtex_brain_putamen_basal |
| rs10835196 | 11 | 27601335 | BDNF | ENSG00000176697.14 | 3.637E-6 | 0.03 | 142270 | gtex_brain_putamen_basal |
| rs12786828 | 11 | 27602642 | BDNF | ENSG00000176697.14 | 3.638E-6 | 0.03 | 140963 | gtex_brain_putamen_basal |
| rs2029364 | 11 | 27605728 | BDNF | ENSG00000176697.14 | 3.637E-6 | 0.03 | 137877 | gtex_brain_putamen_basal |
| rs2029363 | 11 | 27606044 | BDNF | ENSG00000176697.14 | 3.642E-6 | 0.03 | 137561 | gtex_brain_putamen_basal |
| rs2029362 | 11 | 27606133 | BDNF | ENSG00000176697.14 | 3.642E-6 | 0.03 | 137472 | gtex_brain_putamen_basal |
| rs1948186 | 11 | 27610574 | BDNF | ENSG00000176697.14 | 3.66E-6 | 0.03 | 133031 | gtex_brain_putamen_basal |
| rs11030058 | 11 | 27616499 | BDNF | ENSG00000176697.14 | 4.021E-6 | 0.03 | 127106 | gtex_brain_putamen_basal |
| rs11030059 | 11 | 27616617 | BDNF | ENSG00000176697.14 | 3.974E-6 | 0.03 | 126988 | gtex_brain_putamen_basal |
| rs10835199 | 11 | 27617034 | BDNF | ENSG00000176697.14 | 4.112E-6 | 0.03 | 126571 | gtex_brain_putamen_basal |
| rs10835200 | 11 | 27617074 | BDNF | ENSG00000176697.14 | 4.114E-6 | 0.03 | 126531 | gtex_brain_putamen_basal |
| rs11030060 | 11 | 27617566 | BDNF | ENSG00000176697.14 | 3.968E-6 | 0.03 | 126039 | gtex_brain_putamen_basal |
| rs11030062 | 11 | 27617626 | BDNF | ENSG00000176697.14 | 3.998E-6 | 0.03 | 125979 | gtex_brain_putamen_basal |
| rs11030064 | 11 | 27618016 | BDNF | ENSG00000176697.14 | 4.227E-6 | 0.03 | 125589 | gtex_brain_putamen_basal |
| rs11030065 | 11 | 27618108 | BDNF | ENSG00000176697.14 | 4.232E-6 | 0.03 | 125497 | gtex_brain_putamen_basal |
| rs10835202 | 11 | 27618555 | BDNF | ENSG00000176697.14 | 4.243E-6 | 0.03 | 125050 | gtex_brain_putamen_basal |
| rs7937150 | 11 | 27620360 | BDNF | ENSG00000176697.14 | 4.236E-6 | 0.03 | 123245 | gtex_brain_putamen_basal |
| rs7937485 | 11 | 27620473 | BDNF | ENSG00000176697.14 | 4.236E-6 | 0.03 | 123132 | gtex_brain_putamen_basal |
| rs12798439 | 11 | 27621960 | BDNF | ENSG00000176697.14 | 4.236E-6 | 0.03 | 121645 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
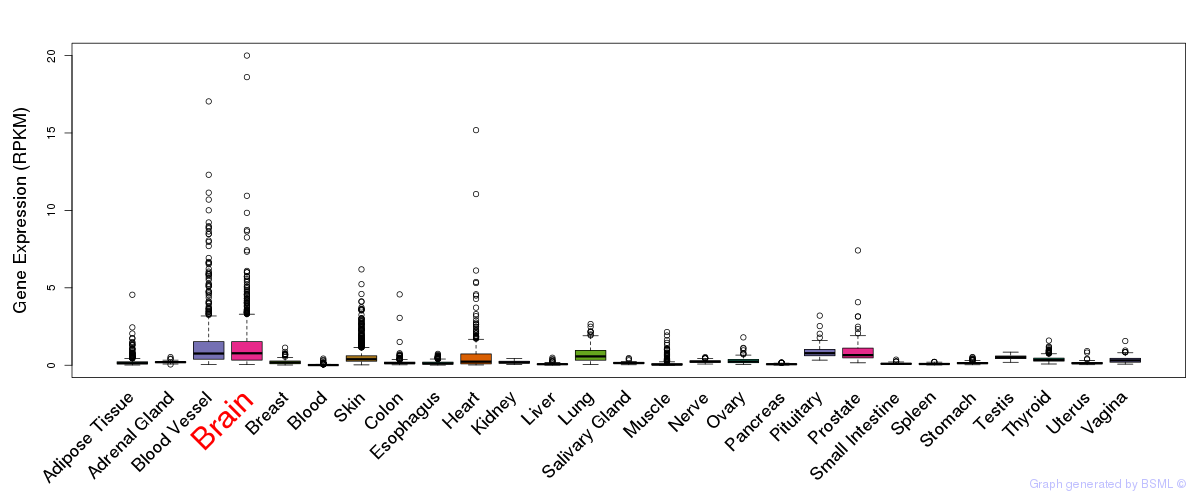
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0008083 | growth factor activity | IEA | - | |
| GO:0008083 | growth factor activity | TAS | 2236018 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007412 | axon target recognition | IEA | axon (GO term level: 13) | - |
| GO:0007411 | axon guidance | IEA | axon (GO term level: 13) | - |
| GO:0007406 | negative regulation of neuroblast proliferation | IEA | neurogenesis (GO term level: 10) | - |
| GO:0008038 | neuron recognition | IEA | neuron (GO term level: 4) | - |
| GO:0014047 | glutamate secretion | IEA | glutamate, Neurotransmitter (GO term level: 7) | - |
| GO:0048167 | regulation of synaptic plasticity | IEA | Synap (GO term level: 8) | - |
| GO:0042490 | mechanoreceptor differentiation | IEA | neuron (GO term level: 9) | - |
| GO:0045666 | positive regulation of neuron differentiation | IEA | neuron (GO term level: 10) | - |
| GO:0043524 | negative regulation of neuron apoptosis | IEA | neuron (GO term level: 9) | - |
| GO:0016358 | dendrite development | IEA | neurite, dendrite (GO term level: 11) | - |
| GO:0001657 | ureteric bud development | IEA | - | |
| GO:0007631 | feeding behavior | IEA | - | |
| GO:0007611 | learning or memory | IEA | - | |
| GO:0006916 | anti-apoptosis | IEA | - | |
| GO:0042493 | response to drug | IEA | - | |
| GO:0042596 | fear response | IEA | - | |
| GO:0021675 | nerve development | IEA | - | |
| GO:0019222 | regulation of metabolic process | IEA | - | |
| GO:0046668 | regulation of retinal cell programmed cell death | IEA | - | |
| GO:0048839 | inner ear development | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0016023 | cytoplasmic membrane-bounded vesicle | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG NEUROTROPHIN SIGNALING PATHWAY | 126 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG HUNTINGTONS DISEASE | 185 | 109 | All SZGR 2.0 genes in this pathway |
| PID SHP2 PATHWAY | 58 | 46 | All SZGR 2.0 genes in this pathway |
| PID AJDISS 2PATHWAY | 48 | 38 | All SZGR 2.0 genes in this pathway |
| PID P75 NTR PATHWAY | 69 | 51 | All SZGR 2.0 genes in this pathway |
| PID TRKR PATHWAY | 62 | 48 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL UP | 285 | 181 | All SZGR 2.0 genes in this pathway |
| WILCOX RESPONSE TO PROGESTERONE DN | 66 | 44 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER BASAL VS MESENCHYMAL DN | 50 | 36 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS UP | 290 | 177 | All SZGR 2.0 genes in this pathway |
| RODRIGUES NTN1 TARGETS UP | 17 | 11 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 8D UP | 157 | 91 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN | 459 | 276 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 12HR DN | 209 | 122 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 XPCS DN | 88 | 71 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER DN | 514 | 319 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| INGRAM SHH TARGETS UP | 127 | 79 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 120 HELA | 69 | 47 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO IKK INHIBITOR AND TNF DN | 103 | 64 | All SZGR 2.0 genes in this pathway |
| KANG IMMORTALIZED BY TERT UP | 89 | 61 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS UP | 214 | 133 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| MODY HIPPOCAMPUS POSTNATAL | 63 | 50 | All SZGR 2.0 genes in this pathway |
| WESTON VEGFA TARGETS 6HR | 59 | 38 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 10HR DN | 56 | 37 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS DN | 371 | 218 | All SZGR 2.0 genes in this pathway |
| MCCLUNG DELTA FOSB TARGETS 2WK | 48 | 36 | All SZGR 2.0 genes in this pathway |
| WESTON VEGFA TARGETS 12HR | 35 | 23 | All SZGR 2.0 genes in this pathway |
| WESTON VEGFA TARGETS | 108 | 71 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 8 | 86 | 57 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D2 | 41 | 30 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE DN | 261 | 183 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| MCCLUNG CREB1 TARGETS UP | 100 | 72 | All SZGR 2.0 genes in this pathway |
| LEE AGING CEREBELLUM UP | 84 | 58 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| LOPES METHYLATED IN COLON CANCER DN | 28 | 26 | All SZGR 2.0 genes in this pathway |
| SANSOM APC MYC TARGETS | 217 | 138 | All SZGR 2.0 genes in this pathway |
| MCCABE BOUND BY HOXC6 | 469 | 239 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN UP | 439 | 257 | All SZGR 2.0 genes in this pathway |
| TOYOTA TARGETS OF MIR34B AND MIR34C | 463 | 262 | All SZGR 2.0 genes in this pathway |
| MISHRA CARCINOMA ASSOCIATED FIBROBLAST DN | 24 | 13 | All SZGR 2.0 genes in this pathway |
| LIN NPAS4 TARGETS DN | 68 | 48 | All SZGR 2.0 genes in this pathway |
| HELLEBREKERS SILENCED DURING TUMOR ANGIOGENESIS | 80 | 56 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP | 395 | 249 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING DN | 225 | 124 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN | 253 | 192 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| HOELZEL NF1 TARGETS DN | 115 | 73 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS DN | 315 | 215 | All SZGR 2.0 genes in this pathway |
| WINZEN DEGRADED VIA KHSRP | 100 | 70 | All SZGR 2.0 genes in this pathway |
| IKEDA MIR1 TARGETS UP | 53 | 39 | All SZGR 2.0 genes in this pathway |
| IKEDA MIR30 TARGETS UP | 116 | 87 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE VIA P38 PARTIAL | 160 | 106 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
| NABA SECRETED FACTORS | 344 | 197 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-1/206 | 220 | 226 | m8 | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA | ||||
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA | ||||
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-10 | 65 | 72 | 1A,m8 | hsa-miR-10a | UACCCUGUAGAUCCGAAUUUGUG |
| hsa-miR-10b | UACCCUGUAGAACCGAAUUUGU | ||||
| miR-103/107 | 299 | 305 | m8 | hsa-miR-103brain | AGCAGCAUUGUACAGGGCUAUGA |
| hsa-miR-107brain | AGCAGCAUUGUACAGGGCUAUCA | ||||
| miR-15/16/195/424/497 | 300 | 306 | m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-155 | 2797 | 2803 | 1A | hsa-miR-155 | UUAAUGCUAAUCGUGAUAGGGG |
| miR-182 | 252 | 258 | m8 | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-191 | 393 | 399 | m8 | hsa-miR-191brain | CAACGGAAUCCCAAAAGCAGCU |
| miR-210 | 196 | 202 | 1A | hsa-miR-210 | CUGUGCGUGUGACAGCGGCUGA |
| miR-30-5p | 397 | 403 | 1A | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-365 | 2796 | 2802 | 1A | hsa-miR-365 | UAAUGCCCCUAAAAAUCCUUAU |
| miR-381 | 11 | 17 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU | ||||
| miR-382 | 199 | 206 | 1A,m8 | hsa-miR-382brain | GAAGUUGUUCGUGGUGGAUUCG |
| miR-409-3p | 389 | 395 | m8 | hsa-miR-409-3p | CGAAUGUUGCUCGGUGAACCCCU |
| miR-410 | 2768 | 2774 | 1A | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| miR-495 | 552 | 558 | m8 | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
| hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU | ||||
| hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU | ||||
| hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU | ||||
| hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU | ||||
| miR-496 | 2727 | 2734 | 1A,m8 | hsa-miR-496 | AUUACAUGGCCAAUCUC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.