Gene Page: MCM3
Summary ?
| GeneID | 4172 |
| Symbol | MCM3 |
| Synonyms | HCC5|P1-MCM3|P1.h|RLFB |
| Description | minichromosome maintenance complex component 3 |
| Reference | MIM:602693|HGNC:HGNC:6945|Ensembl:ENSG00000112118|HPRD:04072|Vega:OTTHUMG00000014844 |
| Gene type | protein-coding |
| Map location | 6p12 |
| Pascal p-value | 0.194 |
| Sherlock p-value | 0.277 |
| Fetal beta | 0.531 |
| DMG | 2 (# studies) |
| eGene | Caudate basal ganglia Cortex Frontal Cortex BA9 Nucleus accumbens basal ganglia Putamen basal ganglia |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Montano_2016 | Genome-wide DNA methylation analysis | This dataset includes 172 replicated associations between CpGs with schizophrenia. | 2 |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 2 |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.04433 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0069 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg07333545 | 6 | 52170606 | MCM3 | 1.44E-5 | -0.011 | 0.052 | DMG:Montano_2016 |
| cg27638713 | 6 | 52149159 | MCM3 | -0.024 | 0.4 | DMG:Nishioka_2013 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6933615 | 6 | 52264514 | MCM3 | ENSG00000112118.13 | 2.722E-7 | 0 | -114835 | gtex_brain_putamen_basal |
| rs112358192 | 6 | 52277217 | MCM3 | ENSG00000112118.13 | 2.304E-7 | 0 | -127538 | gtex_brain_putamen_basal |
| rs9474218 | 6 | 52277984 | MCM3 | ENSG00000112118.13 | 2.722E-7 | 0 | -128305 | gtex_brain_putamen_basal |
| rs58685840 | 6 | 52285060 | MCM3 | ENSG00000112118.13 | 5.573E-8 | 0 | -135381 | gtex_brain_putamen_basal |
| rs546247 | 6 | 52292881 | MCM3 | ENSG00000112118.13 | 1.515E-8 | 0 | -143202 | gtex_brain_putamen_basal |
| rs67642847 | 6 | 52301785 | MCM3 | ENSG00000112118.13 | 1.516E-8 | 0 | -152106 | gtex_brain_putamen_basal |
| rs67736214 | 6 | 52302042 | MCM3 | ENSG00000112118.13 | 1.516E-8 | 0 | -152363 | gtex_brain_putamen_basal |
| rs539295 | 6 | 52303048 | MCM3 | ENSG00000112118.13 | 1.516E-8 | 0 | -153369 | gtex_brain_putamen_basal |
| rs553329 | 6 | 52306670 | MCM3 | ENSG00000112118.13 | 5.881E-9 | 0 | -156991 | gtex_brain_putamen_basal |
| rs631603 | 6 | 52314213 | MCM3 | ENSG00000112118.13 | 1.516E-8 | 0 | -164534 | gtex_brain_putamen_basal |
| rs642679 | 6 | 52314384 | MCM3 | ENSG00000112118.13 | 1.516E-8 | 0 | -164705 | gtex_brain_putamen_basal |
| rs111837113 | 6 | 52315259 | MCM3 | ENSG00000112118.13 | 1.516E-8 | 0 | -165580 | gtex_brain_putamen_basal |
| rs59160834 | 6 | 52324249 | MCM3 | ENSG00000112118.13 | 1.516E-8 | 0 | -174570 | gtex_brain_putamen_basal |
| rs2104333 | 6 | 52324496 | MCM3 | ENSG00000112118.13 | 1.516E-8 | 0 | -174817 | gtex_brain_putamen_basal |
| rs60675996 | 6 | 52324733 | MCM3 | ENSG00000112118.13 | 1.504E-8 | 0 | -175054 | gtex_brain_putamen_basal |
| rs16882561 | 6 | 52329510 | MCM3 | ENSG00000112118.13 | 1.516E-8 | 0 | -179831 | gtex_brain_putamen_basal |
| rs66608904 | 6 | 52331777 | MCM3 | ENSG00000112118.13 | 1.516E-8 | 0 | -182098 | gtex_brain_putamen_basal |
| rs112649755 | 6 | 52333837 | MCM3 | ENSG00000112118.13 | 1.516E-8 | 0 | -184158 | gtex_brain_putamen_basal |
| rs111743208 | 6 | 52336926 | MCM3 | ENSG00000112118.13 | 1.516E-8 | 0 | -187247 | gtex_brain_putamen_basal |
| rs616248 | 6 | 52338522 | MCM3 | ENSG00000112118.13 | 1.516E-8 | 0 | -188843 | gtex_brain_putamen_basal |
| rs1266787 | 6 | 52343899 | MCM3 | ENSG00000112118.13 | 1.516E-8 | 0 | -194220 | gtex_brain_putamen_basal |
| rs515432 | 6 | 52344662 | MCM3 | ENSG00000112118.13 | 1.516E-8 | 0 | -194983 | gtex_brain_putamen_basal |
| rs1266789 | 6 | 52346506 | MCM3 | ENSG00000112118.13 | 1.516E-8 | 0 | -196827 | gtex_brain_putamen_basal |
| rs55934038 | 6 | 52348455 | MCM3 | ENSG00000112118.13 | 1.516E-8 | 0 | -198776 | gtex_brain_putamen_basal |
| rs55953467 | 6 | 52356259 | MCM3 | ENSG00000112118.13 | 1.516E-8 | 0 | -206580 | gtex_brain_putamen_basal |
| rs72925235 | 6 | 52361027 | MCM3 | ENSG00000112118.13 | 1.519E-8 | 0 | -211348 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
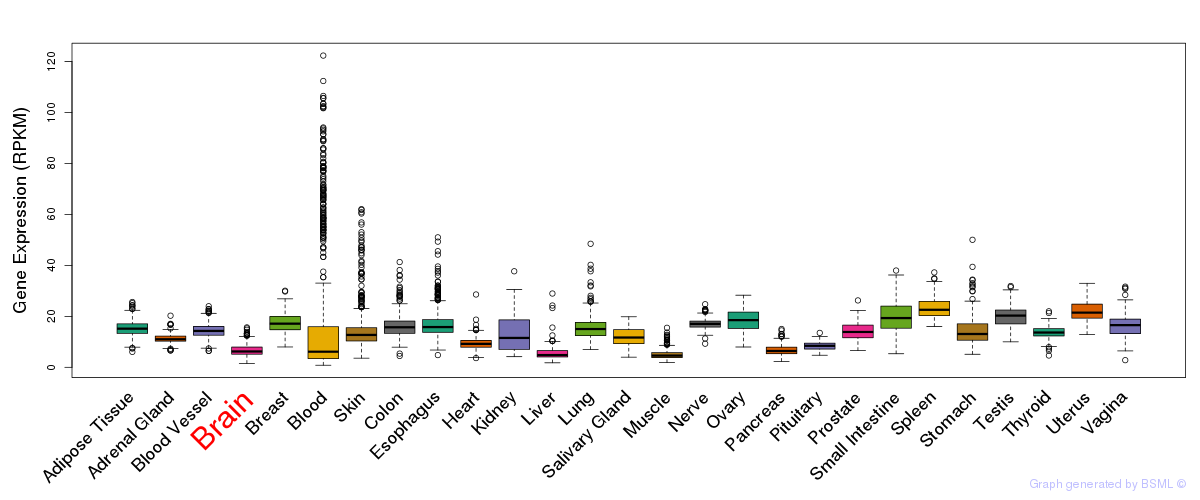
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003677 | DNA binding | IEA | - | |
| GO:0003677 | DNA binding | TAS | 1549468 | |
| GO:0005515 | protein binding | IPI | 12364596 |15232106 |15654075 |17353931 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0017111 | nucleoside-triphosphatase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0006270 | DNA replication initiation | TAS | 1549468 | |
| GO:0006260 | DNA replication | EXP | 12791985 | |
| GO:0006260 | DNA replication | IEA | - | |
| GO:0007049 | cell cycle | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IDA | 18029348 | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005654 | nucleoplasm | EXP | 10436018 |10846177 |11095689 |11931757 |12045100 |15226314 |15707391 | |
| GO:0005658 | alpha DNA polymerase:primase complex | TAS | 1549468 | |
| GO:0005730 | nucleolus | IDA | 18029348 | |
| GO:0005737 | cytoplasm | IDA | 18029348 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CCDC5 | FLJ21094 | FLJ40084 | HEI-C | HsT1461 | coiled-coil domain containing 5 (spindle associated) | - | HPRD | 15232106 |
| CDC45L | CDC45 | CDC45L2 | PORC-PI-1 | CDC45 cell division cycle 45-like (S. cerevisiae) | Two-hybrid | BioGRID | 12614612 |
| CDC6 | CDC18L | HsCDC18 | HsCDC6 | cell division cycle 6 homolog (S. cerevisiae) | - | HPRD,BioGRID | 10464337 |11046155 |
| CDC7 | CDC7L1 | HsCDC7 | Hsk1 | MGC117361 | MGC126237 | MGC126238 | huCDC7 | cell division cycle 7 homolog (S. cerevisiae) | - | HPRD,BioGRID | 9250678 |
| CDC7 | CDC7L1 | HsCDC7 | Hsk1 | MGC117361 | MGC126237 | MGC126238 | huCDC7 | cell division cycle 7 homolog (S. cerevisiae) | - | HPRD | 9250678 |15232106 |
| CDK2 | p33(CDK2) | cyclin-dependent kinase 2 | - | HPRD | 15232106 |
| DBF4 | ASK | CHIF | DBF4A | ZDBF1 | DBF4 homolog (S. cerevisiae) | - | HPRD,BioGRID | 12614612 |
| ENSA | MGC4319 | MGC78563 | MGC8394 | endosulfine alpha | Affinity Capture-MS | BioGRID | 17353931 |
| MCM10 | CNA43 | DNA43 | MGC126776 | PRO2249 | minichromosome maintenance complex component 10 | - | HPRD,BioGRID | 11095689 |15232106 |
| MCM2 | BM28 | CCNL1 | CDCL1 | D3S3194 | KIAA0030 | MGC10606 | MITOTIN | cdc19 | minichromosome maintenance complex component 2 | - | HPRD,BioGRID | 12614612 |
| MCM2 | BM28 | CCNL1 | CDCL1 | D3S3194 | KIAA0030 | MGC10606 | MITOTIN | cdc19 | minichromosome maintenance complex component 2 | Mcm2 interacts with Mcm3. | BIND | 15236977 |
| MCM2 | BM28 | CCNL1 | CDCL1 | D3S3194 | KIAA0030 | MGC10606 | MITOTIN | cdc19 | minichromosome maintenance complex component 2 | MCM2 interacts with MCM3. | BIND | 15236977 |
| MCM3AP | FLJ44336 | FLJ45306 | GANP | KIAA0572 | MAP80 | minichromosome maintenance complex component 3 associated protein | - | HPRD | 9712829 |10733502 |11024281 |12226073 |
| MCM3AP | FLJ44336 | FLJ45306 | GANP | KIAA0572 | MAP80 | minichromosome maintenance complex component 3 associated protein | Affinity Capture-Western Two-hybrid | BioGRID | 9712829 |10733502 |12226073 |
| MCM5 | CDC46 | MGC5315 | P1-CDC46 | minichromosome maintenance complex component 5 | - | HPRD,BioGRID | 11248027 |12614612 |
| MCM5 | CDC46 | MGC5315 | P1-CDC46 | minichromosome maintenance complex component 5 | MCM3 interacts with MCM5. | BIND | 15236977 |
| MCM5 | CDC46 | MGC5315 | P1-CDC46 | minichromosome maintenance complex component 5 | Mcm3 interacts with Mcm5. | BIND | 15236977 |
| MCM7 | CDABP0042 | CDC47 | MCM2 | P1.1-MCM3 | P1CDC47 | P85MCM | PNAS-146 | minichromosome maintenance complex component 7 | MCM3 interacts with MCM7. | BIND | 15236977 |
| MCM7 | CDABP0042 | CDC47 | MCM2 | P1.1-MCM3 | P1CDC47 | P85MCM | PNAS-146 | minichromosome maintenance complex component 7 | - | HPRD | 7601140 |8631321 |10464337 |
| MCM7 | CDABP0042 | CDC47 | MCM2 | P1.1-MCM3 | P1CDC47 | P85MCM | PNAS-146 | minichromosome maintenance complex component 7 | Mcm3 interacts with Mcm7. | BIND | 15236977 |
| MCM7 | CDABP0042 | CDC47 | MCM2 | P1.1-MCM3 | P1CDC47 | P85MCM | PNAS-146 | minichromosome maintenance complex component 7 | Affinity Capture-Western Co-fractionation Two-hybrid | BioGRID | 9099751 |10464337 |11779870 |12614612 |
| ORC4L | ORC4 | ORC4P | origin recognition complex, subunit 4-like (yeast) | Two-hybrid | BioGRID | 12614612 |
| ORC5L | ORC5 | ORC5P | ORC5T | origin recognition complex, subunit 5-like (yeast) | Two-hybrid | BioGRID | 12614612 |
| PAK2 | PAK65 | PAKgamma | p21 protein (Cdc42/Rac)-activated kinase 2 | Affinity Capture-MS | BioGRID | 17353931 |
| PNKP | PNK | polynucleotide kinase 3'-phosphatase | Affinity Capture-MS | BioGRID | 17353931 |
| PTP4A3 | PRL-3 | PRL-R | PRL3 | protein tyrosine phosphatase type IVA, member 3 | Affinity Capture-MS | BioGRID | 17353931 |
| RPA2 | REPA2 | RPA32 | replication protein A2, 32kDa | Two-hybrid | BioGRID | 12614612 |
| SNCG | BCSG1 | SR | synuclein, gamma (breast cancer-specific protein 1) | Affinity Capture-MS | BioGRID | 17353931 |
| STAT1 | DKFZp686B04100 | ISGF-3 | STAT91 | signal transducer and activator of transcription 1, 91kDa | - | HPRD | 11248027 |
| WDR8 | FLJ20430 | MGC99569 | WD repeat domain 8 | Affinity Capture-MS | BioGRID | 17353931 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG DNA REPLICATION | 36 | 21 | All SZGR 2.0 genes in this pathway |
| KEGG CELL CYCLE | 128 | 84 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MCM PATHWAY | 18 | 8 | All SZGR 2.0 genes in this pathway |
| PID E2F PATHWAY | 74 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | 31 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME ORC1 REMOVAL FROM CHROMATIN | 67 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE CHECKPOINTS | 124 | 70 | All SZGR 2.0 genes in this pathway |
| REACTOME M G1 TRANSITION | 81 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME G1 S TRANSITION | 112 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS OF DNA | 92 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC G1 G1 S PHASES | 137 | 79 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC M M G1 PHASES | 172 | 98 | All SZGR 2.0 genes in this pathway |
| REACTOME ASSEMBLY OF THE PRE REPLICATIVE COMPLEX | 65 | 36 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPLICATION | 192 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | 38 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME UNWINDING OF DNA | 11 | 6 | All SZGR 2.0 genes in this pathway |
| REACTOME G2 M CHECKPOINTS | 45 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME S PHASE | 109 | 66 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA STRAND ELONGATION | 30 | 18 | All SZGR 2.0 genes in this pathway |
| PYEON HPV POSITIVE TUMORS UP | 98 | 47 | All SZGR 2.0 genes in this pathway |
| SOTIRIOU BREAST CANCER GRADE 1 VS 3 UP | 151 | 84 | All SZGR 2.0 genes in this pathway |
| PUIFFE INVASION INHIBITED BY ASCITES UP | 82 | 51 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS UP | 290 | 177 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER 20Q13 AMPLIFICATION DN | 180 | 101 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| PROVENZANI METASTASIS UP | 194 | 112 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF DN | 228 | 137 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| JOHANSSON GLIOMAGENESIS BY PDGFB UP | 58 | 44 | All SZGR 2.0 genes in this pathway |
| MARKS HDAC TARGETS UP | 23 | 15 | All SZGR 2.0 genes in this pathway |
| KONG E2F3 TARGETS | 97 | 58 | All SZGR 2.0 genes in this pathway |
| EGUCHI CELL CYCLE RB1 TARGETS | 23 | 13 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION | 77 | 51 | All SZGR 2.0 genes in this pathway |
| LI LUNG CANCER | 41 | 30 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| LIANG HEMATOPOIESIS STEM CELL NUMBER SMALL VS HUGE DN | 33 | 22 | All SZGR 2.0 genes in this pathway |
| PUJANA BREAST CANCER LIT INT NETWORK | 101 | 73 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN MELANOMA RELAPSE UP | 61 | 25 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| CAFFAREL RESPONSE TO THC DN | 31 | 23 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY UP | 236 | 139 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED MODERATELY VS POORLY DN | 15 | 8 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION UP | 69 | 55 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES 1 | 379 | 235 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| BENPORATH PROLIFERATION | 147 | 80 | All SZGR 2.0 genes in this pathway |
| GEORGES CELL CYCLE MIR192 TARGETS | 62 | 46 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| SUNG METASTASIS STROMA DN | 54 | 34 | All SZGR 2.0 genes in this pathway |
| MORI LARGE PRE BII LYMPHOCYTE UP | 86 | 49 | All SZGR 2.0 genes in this pathway |
| MORI IMMATURE B LYMPHOCYTE DN | 90 | 55 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPLICATION GENES | 147 | 87 | All SZGR 2.0 genes in this pathway |
| GOLUB ALL VS AML UP | 24 | 20 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO DN | 200 | 112 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA DN | 289 | 166 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST DN | 309 | 206 | All SZGR 2.0 genes in this pathway |
| REN BOUND BY E2F | 61 | 40 | All SZGR 2.0 genes in this pathway |
| PEART HDAC PROLIFERATION CLUSTER DN | 76 | 57 | All SZGR 2.0 genes in this pathway |
| SASAKI ADULT T CELL LEUKEMIA | 176 | 122 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS UP | 126 | 84 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| HESS TARGETS OF HOXA9 AND MEIS1 UP | 65 | 44 | All SZGR 2.0 genes in this pathway |
| YAGI AML FAB MARKERS | 191 | 131 | All SZGR 2.0 genes in this pathway |
| IGLESIAS E2F TARGETS UP | 151 | 103 | All SZGR 2.0 genes in this pathway |
| KAMMINGA EZH2 TARGETS | 41 | 26 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR | 532 | 309 | All SZGR 2.0 genes in this pathway |
| KAAB FAILED HEART ATRIUM DN | 141 | 99 | All SZGR 2.0 genes in this pathway |
| RHODES CANCER META SIGNATURE | 64 | 47 | All SZGR 2.0 genes in this pathway |
| RHODES UNDIFFERENTIATED CANCER | 69 | 44 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 3 | 101 | 64 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR UP | 178 | 111 | All SZGR 2.0 genes in this pathway |
| SONG TARGETS OF IE86 CMV PROTEIN | 60 | 42 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN DN | 271 | 175 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS PEAK AT 16HR | 40 | 24 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| SARRIO EPITHELIAL MESENCHYMAL TRANSITION UP | 180 | 114 | All SZGR 2.0 genes in this pathway |
| FUJII YBX1 TARGETS DN | 202 | 132 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| TOYOTA TARGETS OF MIR34B AND MIR34C | 463 | 262 | All SZGR 2.0 genes in this pathway |
| ZHENG GLIOBLASTOMA PLASTICITY UP | 250 | 168 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| GRADE COLON AND RECTAL CANCER UP | 285 | 167 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB DN | 342 | 220 | All SZGR 2.0 genes in this pathway |
| MUELLER PLURINET | 299 | 189 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C UP | 47 | 29 | All SZGR 2.0 genes in this pathway |
| FRASOR RESPONSE TO SERM OR FULVESTRANT DN | 50 | 29 | All SZGR 2.0 genes in this pathway |
| GOLDRATH ANTIGEN RESPONSE | 346 | 192 | All SZGR 2.0 genes in this pathway |
| RUIZ TNC TARGETS DN | 142 | 79 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE UP | 212 | 128 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR DN | 277 | 166 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| CROONQUIST IL6 DEPRIVATION DN | 98 | 69 | All SZGR 2.0 genes in this pathway |
| CROONQUIST NRAS SIGNALING DN | 72 | 47 | All SZGR 2.0 genes in this pathway |
| CROONQUIST NRAS VS STROMAL STIMULATION DN | 99 | 65 | All SZGR 2.0 genes in this pathway |
| ISHIDA E2F TARGETS | 53 | 27 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G23 UP | 52 | 35 | All SZGR 2.0 genes in this pathway |
| KOBAYASHI EGFR SIGNALING 24HR DN | 251 | 151 | All SZGR 2.0 genes in this pathway |
| FOURNIER ACINAR DEVELOPMENT LATE 2 | 277 | 172 | All SZGR 2.0 genes in this pathway |
| PYEON CANCER HEAD AND NECK VS CERVICAL UP | 193 | 95 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA UP | 207 | 143 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| WONG EMBRYONIC STEM CELL CORE | 335 | 193 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 12 | 79 | 54 | All SZGR 2.0 genes in this pathway |
| WU APOPTOSIS BY CDKN1A VIA TP53 | 55 | 31 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS LOW SERUM | 100 | 51 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS GROWING | 243 | 155 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 DN | 374 | 217 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 6HR UP | 229 | 149 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR UP | 324 | 193 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 DN | 336 | 211 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL UP | 289 | 184 | All SZGR 2.0 genes in this pathway |
| ZHOU CELL CYCLE GENES IN IR RESPONSE 6HR | 85 | 49 | All SZGR 2.0 genes in this pathway |
| ZHOU CELL CYCLE GENES IN IR RESPONSE 24HR | 128 | 73 | All SZGR 2.0 genes in this pathway |